Journal of Food: Microbiology, Safety & Hygiene
Open Access
ISSN: 2476-2059
ISSN: 2476-2059
Research Article - (2025)Volume 10, Issue 1
Raw beef consumption is an old tradition and a much favored dish by the majority of Ethiopians. However, prevailing unhygienic processing and distribution practices likely contribute to beef contamination leading to foodborne infections. A cross-sectional study was carried out to investigate the occurrence of E. coli O157:H7 and evaluate its antimicrobial resistance profile in slaughterhouses and butcher shops in Bishoftu town, Ethiopia. A total of 352 samples (120 fecal, 92 beef cut, and 140 environmental swabs) were collected. The isolation and identification process was carried out using selective enrichment media, followed by a latex agglutination test. The isolates were tested for their resistance against 13 antimicrobials using the standard disk diffusion method. Out of 352 samples, 14 (3.97%) were found to be positive for E. coli O157:H7 serotype; of which, 28.6% (4/14), 21.4% (3/14), and 50% (7/14) were from fecal, beef, and environmental swab samples respectively. A significant difference in the occurrences of the pathogen was observed among the sources of samples (p<0.05). Results of the antimicrobial susceptibility test revealed high resistance to three commonly used drugs: Tetracycline (100%), erythromycin (92.8%), and ampicillin (64.3%). All of the E. coli O157 isolates were found to be susceptible to azithromycin, cefotaxime, and chloramphenicol. Of the 14 isolates, 12 (85.8%) of them were found to be resistant to three or more classes of antimicrobial agents. E. coli O157:H7 was detected in samples collected from meat and environmental samples implying the health risk of raw and undercooked meat consumption. Results also showed occurrences of multiple antimicrobial-resistant E. coli O157:H7. Therefore, the current study warrants the need of implementing appropriate hygienic measures in slaughterhouses and butcher shops to safeguard public health.
Beef; Contamination, E. coli O157:H7; Drug resistance
Despite the improved technology and hygienic practices at all stages of food production, foodborne diseases are continued to be great public health and well-being concerns of individuals and countries across the world. Especially, developing countries are largely vulnerable to food-borne infections which cost billions of dollars in medical care and social costs. Studies indicated that each year, 1 out of 10 people get ill from microbial food contamination, resulting in 600 million illnesses, 420 000 deaths, and the loss of 33 million healthy years of life globally. It often follows the consumption of contaminated foodstuffs, especially from animal products such as meat from infected animals or carcasses contaminated with pathogenic bacteria including Escherichia coli [1].
E. coli O157:H7, an Entero-Hemorrhagic E. coli (EHEC), is one of the most common causes of foodborne infections in humans. It infects all age groups and the pathogen is noted for its severe consequences following infection, low infective dose, and acid resistance. Depending on the immune status and the general health of the infected individual, and the dose and virulence of the bacteria, infection with E. coli O157:H7 can result in mild diarrhea, severe bloody diarrhea, hemorrhagic colitis, or Hemolytic Uremic Syndrome (HUS) leading to kidney failure. Globally, EHEC O157:H7 causes 2, 801, 000 acute illnesses annually, with an incidence rate of 43.1 cases per 100,000 persons per year. Among those, a total of 10,200 cases of STEC infections occur in Africa with an incidence rate of 1.4 cases per 100,000 people per year [2].
Cattle are the primary reservoirs of E. coli O157:H7 and consumption of beef and beef products are identified as major sources of foodborne transmission. Carcass contamination occurs through skin-to-carcass or fecal-to-carcass transfer of the pathogen during the slaughter process at processing plants. Furthermore, microbial cross-contamination can occur during processing and manipulation, such as dehiding, evisceration, storage, and distribution at slaughterhouses and butcher shops [3].
To increase the production output, the animal production sector in developing countries have been regularly using antimicrobials for therapy, diseases prevention, and growth purpose. Antimicrobials are widely used in cattle for disease prevention and growth promotion. In Ethiopia, different studies have shown multidrug resistance among E. coli O157:H7 of animal food origin. E. coli O157:H7 can be transmitted to humans through contaminated food and water, directly between persons, and through contact with animals or their environment. The use of antimicrobials in food cattle leads to the development of resistance pathogenic E. coli O157:H7 that can reach humans through the beef food chain [4].
E.coli O157:H7 detection can be done by various methods that include culture-based, immunological-based, nucleic acid-based, and biosensors. The value of a latex agglutination test for the rapid presumptive detection of E. coli serotype 0157:H7 was determined by laboratory trials and during an outbreak of hemorrhagic colitis. The latex test was found to be a simple, highly efficient, and reliable test in detecting E. coli 0157:H7 with 100% sensitivity and specificity. It was also found that sorbitol-MacConkey agar cultures were not as useful for food samples as they were for fecal specimens in screening for E. coli 0157:H7, but the use of the latex screen was particularly efficient in this setting [5].
In Ethiopia, animals are commonly slaughtered and dressed under unhygienic conditions in the open air or sub-standard slaughterhouses which compromises the microbiological quality and safety of the meat obtained from the animals. Lack of surveillance of food-borne pathogens, lack of education and training among slaughterhouse and butcher shop workers, and poor hygienic practice of food handlers are major factor contributing to the high risk of exposure of Ethiopians to foodborne pathogens such as EHEC. Furthermore, raw meat is widely consumed in the country and if hygiene and adequate temperature control are not maintained, these may pose a potential risk for the occurrence of foodborne disease because of a widespread tradition of raw meat consumption in the country. There is a need to investigate the possible sources of E. coli O157:H7 in the beef supply chain, quantify risk factors, and evaluate the hygienic performance of slaughterhouses and butcher shops to ensure that prevention and control strategies are appropriate. However, in Ethiopia, it has not yet been well handled to evaluate the standards of slaughterhouses and their environment which could serve as sources of E. coli O157:H7. Therefore, the objective of this study was to investigate the occurrences and antimicrobial resistance profiles of E. coli O157:H7 from slaughterhouses and butcher shops [6].
Study area
The study was conducted in Bishoftu city, located at 9°N latitude and 40°E longitudes at an altitude of 1850 m above sea level in the central highlands of Ethiopia. According to the CSA, the total population of the town is 197,557. Bishoftu is being a resort city that attracts local and international visitors and guests; as a result, the booming of hotels and restaurants, and business is believed to increase the number of butcher shops. In Bishoftu town, there are three export abattoirs, two medium-scale slaughterhouses (one municipal and one private) and 131 officially registered butcher shops (Figure 1) [7].
Figure 1: Map of the study area.
Study design and sampling methods
A cross-sectional study type was carried out from November 2021 to May 2022 at two slaughterhouses (Bishoftu municipal and privately owned), and 92 butcher shops. Sampling was carried out once a week, where 10 cattle were selected by using a simple random sampling method. Fecal contents of the animals, carcass swabs, and swab samples from the environment (knives, hooks, hand swabs, water, and wastewater) were collected. Fecal samples were collected from each selected animal directly from the rectum using rectal gloves in the lairage. Carcass swab samples were collected after skinning and evisceration. A simple random sampling technique was employed to select 92 butcher shops out of 131, from which beef cuts (at least 25 g) and parallel pooled swab samples of butcher’s knife, hand, and cutting board was collected [8].
Sample size determination and sample collection and transportation
The sample size was determined by the formula and the prevalence of E. coli O157:H7 in cattle and carcass samples was 7.1% and 6.3%, respectively. Therefore, using the 7.1% and 6.3% expected prevalence, 95% confidence interval, and 5% marginal error, the number of cattle and butcher shops was estimated to be 102 and 91, respectively. However, the sample size was increased to compensate for any sample losses as well as to minimize the margin of error. Hence, a total of 120 fecal samples were collected using a sterile arm-length glove from the abattoirs in sterile and leak-proof universal tubes. Rectal fecal sample collections were made. Moreover, a total of 92 beef samples from 92 butcher shops were collected. At least 25 g of beef was taken from the exterior of the carcass (fat tissue) and the surface of lean beef using sterile scalpels and forceps and put into sterile, separately labeled plastic bags. Scalpel and forceps were cleaned with pieces of gauze dipped in 70% ethanol after each sampling to minimize cross-contaminations [9].
Pooled carcass swabs (n=12) were collected from four different sites of the carcass (thorax, brisket, flank, and crutc, one site covering 100 cm2 by placing a sterile template (10 cm × 10 cm) on a carcass. For each sampling area, a sterile cotton-tipped swab (2 cm × 3 cm) fitted with a shaft was moistened in approximately 10 ml of buffered peptone water (Oxoid, Hampshire, England), and was rubbed first horizontally and then vertically several times across the carcass surface. On the completion of the rubbing process, the shaft was broken by pressing it against the inner wall of the test tube and disposed of leaving the cotton swab in the test tube. The four swabs were put into one screwcapped test tube containing 10 ml of sterile buffered peptone water and transported to the Microbiology laboratory of the College of Veterinary Medicine and Agriculture of Addis Ababa University [10].
Environmental samples collection
A total of 128 swab samples (36 from the two slaughterhouses and 92 from butcher shops) were collected. Swab samples collected from the slaughterhouses were the knife, rasp, axe, and hook (n=12) and hand swabs (n=12), and tap water (n=12). Similarly, butcher’s hand swabs, knives, and chopping boards were collected during the operation at the butcher shops. Environmental swab samples were taken from meat contact surfaces of 15 cm2-20 cm2 using a sterile cotton swab moistened in buffered peptone water. Swab samples were immersed in a test tube containing sterile buffer peptone water. In addition, slaughterhouse wastewater samples (50 ml) in the drainage line were collected at every single visit to the slaughterhouses. All samples were labeled legibly with a permanent marker identifying the type/source of the sample, the date of sampling, and the code of the slaughterhouses/butcher shop. The samples were transported in an ice box containing ice packs to the microbiology laboratory of the college of veterinary medicine and agriculture of Addis Ababa university [11].
Isolation of E. coli O157:H7
Twenty-five grams of each fecal and beef cut sample was aseptically transferred into a stomacher bag containing 225 mL of modified tryptone soya broth supplemented with 20 mg/L Novobiocin, homogenized using a stomacher blender (Stomacher 400, Seward Medical, England) for 1 min at normal speed (200 rpm) and incubated at 41.5°C for 6 hours. Similarly, all swab samples from the slaughterhouses and butcher shops were homogenized in 9 mL of mTSBn and incubated at 37°C for 24 hrs [12].
The enriched samples were streaked onto MacConkey agar plates for primary isolation and incubated at 37°C for 24 hrs. Following incubation, the plates were observed for the growth of pink colonies (lactose fermenter). A single, isolated colony was then picked and sub-cultured on Eosin Methylene Blue (EMB) agar for 24 hrs at 37°C for the formation of a metallic sheen. Suspected colonies of E. coli (pinkish color appearance on MacConkey agar and metallic sheen on EMB) were then subcultured onto nutrient agar (Oxoid, England) at 37°C for 24 hrs. From nutrient agar, relevant biochemical tests that included indole, methyl red, Voges-Proskauer reaction, citrate utilization (IMViC) tests, and H2S production tests were performed. A citrate utilization test was also done using Simon’s citrate agar. The test reagents used were Kovac’s reagent for the indole test, methyl red for the methyl red test, and alpha-naphthol and 40% KOH chemicals for Voges-Proskauer reaction tests. The H2S production and citrate utilization test results were observed and interpreted [13].
The bacterium that was confirmed as E. coli was sub-cultured onto Sorbitol MacConkey agar (SMAC) supplemented with 0.05 mg/l Cefixime-2.5 mg/l potassium tellurite, and plates were incubated at 35°C for 20 to 22 hrs. E. coli O157:H7 does not ferment sorbitol and, thus, produces slightly transparent colorless colonies. Then, up to six colorless colonies (non- Sorbitol fermenters) on SMAC agar were picked and subcultured onto nutrient agar slants and incubated at 37°C for 24 hrs for a further confirmatory test [14].
Identification of E. coli O157:H7
Identification and confirmation of non-sorbitol fermenting E. coli O157:H7 were done by latex agglutination test using a latex kit. The latex kit consists of four components: Latex test reagent, latex control reagent, positive controls, and negative controls. The test reagent contains blue latex particles sensitized with a specific antibody against the E. coli O157:H7 antigen and the control reagent consists of latex particles sensitized with rabbit globulin. The positive controls are suspensions of inactivated E. coli O157:H7 cells, whereas the negative controls are suspensions of inactivated non-specific E. coli cells. The test was performed according to the manufacturer's instructions. The latex kit was first checked for its performance by using the control suspensions in the kit, the test was continued after the positive control reacts with the test latex showing a positive result. Briefly, one drop of 0.85% saline water and latex test were dispensed into the reaction card separately. Using a sterile wire loop, a few presumptive colonies of E. coli O157 were taken and emulsified into the saline water on the latex card, then slowly mixed with the test latex and checked for agglutination within 1 minute. A result was positive if agglutination of the latex particles occurred within 1 minute. The negative result was obtained if no agglutination occurred and a smooth blue suspension remained after 60 seconds in the test area. Testpositive isolates were stoked in glycerol using cryovials for further antimicrobial resistance determination [15].
Antimicrobial susceptibility testing
The antimicrobial resistance test was carried out by using the standard agar disc diffusion technique for 13 antimicrobial agents which are in regular use for ruminants, potential public health importance, and recommendations from the guideline of antimicrobial susceptibility test in; Ampicillin (AMP, 10 μg), Azithromycin (AZM, 15 μg), Cefotaxime (CTX, 30 μg), Ceftazidime (CTZ, 30 μg), Chloramphenicol (CHL, 30 μg), Ciprofloxacin (CIP, 5 μg), Colistin sulfate (CT, 10 μg), Erythromycin (ERY, 15 μg), Gentamicin (GEN, 10 μg), Kanamycin (KAN, 30 μg), Nalidixic Acid (NA, 30 μg), Sulfamethoxazole (SXT, 25 μg), and Tetracycline (TET, 30 μg).
Pure colonies of Escherichia coli O157:H7 isolates were transferred into a test tube of 5 ml tryptone soya broth and incubated at 37°C for 6 hours. The turbidity of suspension broth was adjusted using sterile saline solution, or more colonies were added to obtain turbidity that is usually comparable with that of 0.5 McFarland standards. A sterile cotton swab was immersed into the suspension and rotated against the side of the tube to remove the excess fluid and then swabbed uniformly on the surface of already prepared Mueller-Hinton agar (Oxoid,) plates. As soon as the plates dried, antimicrobial discs were placed on the inoculated plates using sterile forceps and incubated at 37°C for 24 hrs. The results were interpreted as resistance, intermediate, or susceptible after the zone of inhibition of the strain was appreciated.
Data management and analysis
The collected data were entered into Microsoft Excel and checked before analysis. The data were analyzed using.
Descriptive statistics were used to summarize the results. The significance of the association between E. coli O157 isolates and sample source, and type of sample was assessed using univariate logistic regression. Odds ratio and 95% confidence intervals were used to measure the strength of associations. A p-value of less than 0.05 was considered significant.
Prevalence of E. coli O157:H7
Table 1 shows a summary of E. coli O157:H7 detections by sample source and type. Accordingly, E. coli were detected in 69 out of 352 (19.6%, 95% CI: 15.6-24.1), of which E. coli O157:H7 strains were detected in 14 (3.97%, 95% CI 2.2-6.6) samples. E. coli O157:H7 was more prevalent in slaughterhouse samples (5.95%, 95% CI: 2.9-10.7) than butcher shops (2.2%, 95% CI: 0.6-5.5) was detected and, Accordingly, E. coli O157:H7 was prevalent in 12.5% of the slaughterhouse samples namely carcasses, carcass contact surfaces, and wastewater samples. E. coli O157:H7 was also detected in 3.33% of fecal samples. Similarly, 3 (3.3%) beef cut samples and 1 (1.1%) carcass contact surfaces sample were positive for E. coli O157:H7 from butcher shops. Higher occurrence of E. coli O157:H7 was observed in the municipal slaughterhouse (8.33%, 95% CI: 3.4-16.4) compared to the private slaughterhouse (3.57%, 95% CI: 0.7-10.1) (Table 1).
| Sample source | Sample type (p*=pooled sample) | No. of samples tested N (%) | No. positive E.coli O157:H7 (%) |
|---|---|---|---|
| Municipal slaughterhouse | Fecal | 60 | 3 (5) |
| Carcass swab (p*) | 6 | 2 (33.3) | |
| Knife swabs (p*) | 6 | 0 | |
| Hand swabs (p*) | 6 | 1 (16.7) | |
| Water/Waste water | 6 | 1 (16.7) | |
| Sub-total | 84 | 7 (8.3) | |
| Private slaughterhouse | Fecal | 60 | 1 (1.7) |
| Carcass swab (p*) | 6 | 1 (16.7) | |
| Knife swabs (p*) | 6 | 1 (16.7) | |
| Hand swabs (p*) | 6 | 0 | |
| Water/Wastewater | 6 | 0 | |
| Sub-total | 84 | 3 (3.57) | |
| Butcher shops | Sub-total of slaughterhouse | 168 | 10 ( 5.95) |
| Beef /Meat | 92 | 3 (3.3) | |
| Butcher swab sample (p*) | 92 | 1 (1.1) | |
| Sub-total | 184 | 4 (2.2) | |
| Total N (%) | 352 | 14 (3.97) |
Table 1: The occurrence of E. coli O157:H7 by sample sources and type.
A univariable logistic regression test was used to assess the associations of E. coli O157:H7 occurrence with sample source and type. Hence, samples from municipal slaughterhouses had a significantly higher prevalence of E. coli O157:H7 compared to butcher shops. The likelihood of E. coli O157:H7 detections from municipal and private slaughterhouses were 4.09 and 1.67 times more than butcher shops. However, no significant association was observed between E. coli O157:H7 detection and sample types. Although there was no significant association with sample type, higher odds of E. coli O157:H7 detection were observed in water (OR=2.6) and swab (OR=1.4) samples than in fecal samples (Table 2).
| Categories | No. of positive | Odd ratio (95% CI) | P=value | |
|---|---|---|---|---|
| Sample source | Butcher house | 4 | Ref | Ref |
| Municipal | 7 | 4.09 (1.20-16.0) | 0.028 | |
| Private | 3 | 1.67 (0.32-7.73) | 0.51 | |
| Sample type | Fecal | 4 | Ref | Ref |
| Beef /Meat | 3 | 0.98 (0.19-4.54) | 0.977 | |
| Swab samples | 6 | 1.43 (0.40-5.70) | 0.59 | |
| Water/Wastewater | 1 | 2.64 (0.13-19.89) | 0.404 | |
Table 2: Association of E. coli O157:H7 with respected sample source and sample type.
Assessment of the hygienic practices in slaughterhouses and butcher shops
The observational survey revealed that besides the lack of training on food safety, the shortage of facilities hindered workers to maintain the minimum acceptable hygienic practices. None of the slaughterhouse and butcher shop workers had proper hand washing using soap and disinfection, and washing and disinfection of their operational tools and floor after each working interval. There was no specifically separate equipment such as cutting boards, knives, or rasps used for both cutting meat and abdominal organs. Moreover, the substandard slaughtering practices were made without necessary precautions in place to avoid cross-contamination.
Antimicrobial resistance profile
Antimicrobial susceptibility testing was performed for all confirmed positive isolates against 13 different antimicrobial agents. Accordingly, E. coli O157:H7 was found to be resistant to tetracycline (100%), erythromycin (92.8%), and ampicillin (64.3%). Furthermore, resistance of 14.3% was observed to ceftazidime, colistin sulfate, kanamycin, nalidixic acid, and sulfamethoxazole. However, none of the isolates was resistant to azithromycin, cefotaxime, and chloramphenicol (Table 3 and Figure 2).
| Antimicrobial disc | Diameter of zone of inhibition (mm) | Resistance profile (n=14) | |||||
|---|---|---|---|---|---|---|---|
| Con. (μg) | R (≤) | I | S (≥) | S (%) | I (%) | R (%) | |
| Ampicillin (AMP) | 10 | 13 | 14-16 | 17 | 3 (21.4) | 2 (14.3) | 9 (64.3) |
| Azithromycin (AZM) | 15 | 12 | - | 13 | 14 (100) | 0 | 0 |
| Cefotaxime (CTX ) | 30 | 22 | 23-25 | 26 | 14 (100) | 0 | 0 |
| Ceftazidime (CTZ) | 30 | 14 | 15-17 | 18 | 11 (78.6) | 1 (7.1) | 2 (14.3) |
| Chloramphenicol (C) | 30 | 12 | 13-17 | 18 | 14 (100) | 0 | 0 |
| Ciprofloxacin (CIP) | 5 | 15 | 16-20 | 21 | 12 (85.8) | 1 (7.1) | 1 (7.1) |
| Colistin sulphate (CT) | 10 | 16 | 17-21 | 21 | 12 (85.8) | 0 | 2 (14.3) |
| Erythromycin (ERY) | 15 | 13 | 14-22 | 23 | 0 | 1 (7.1) | 13 (89.9) |
| Gentamicin (GEN) | 10 | 12 | 13-14 | 15 | 12 (85.8) | 1 (7.1) | 1 (7.1) |
| Kanamycin (KAN) | 30 | 13 | 14-17 | 18 | 11 (78.6) | 1 (7.1) | 2 (14.3) |
| Nalidixic Acid (NA) | 30 | 13 | 14-18 | 19 | 12 (85.7) | 0 | 2 (14.3) |
| Sulfamethoxazole (SXT) | 25 | 10 | 11-15 | 16 | 12 (85.7) | 0 | 2 (14.3) |
| Tetracycline (TTC) | 30 | 11 | 12-14 | 15 | 0 | 0 | 14 (100) |
Note: S=Susceptible, I=Intermediate, R=Resistant
Table 3: Antimicrobial resistance profile of E. coli O157:H7 isolates.
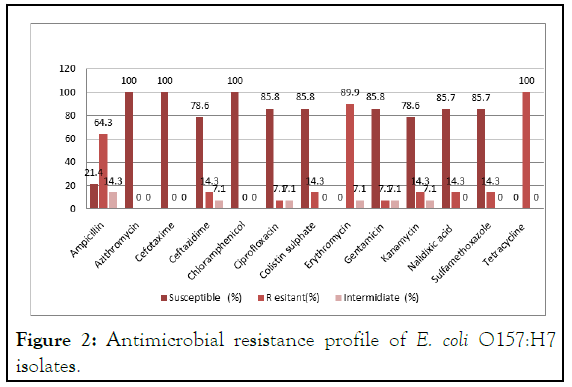
Figure 2: Antimicrobial resistance profile of E. coli O157:H7 isolates.
Multi-drug resistance profiles
This study showed that 12 (85.8%) E. coli O157:H7 isolates were resistant to three or more classes of antimicrobial agents. Multi- Drug Resistance (MDR) profiles of the isolates 5/14 (35.7%), 5/14 (35.7%) and 1 (7.1%), against three, four, and five antimicrobials were registered respectively. Most of the multidrug resistant isolates originated from swabs and fecal samples. A multi-drug resistance profile consisting of six drugs was also documented in 1/14 (7.1%) of the fecal isolate (Table 4).
| Source of isolates | Resistance profile | Antibiotic classes | Sources of MDR | Total MDR | |||
|---|---|---|---|---|---|---|---|
| Beef n=3 | Fecal n=4 | Swab n=6 | Water n=1 | ||||
| Municipal | CT, ERY, TTC | R3 | - | 1 | - | - | 1 |
| Slaughter | AMP, ERY, TTC | R3 | - | - | 1 | 1 | 2 |
| House | AMP, ERY, NA, TTC | R4 | - | 1 | - | - | 1 |
| AMP, ERY, GEN, TTC | R4 | - | - | 1 | - | 1 | |
| AMP, CTX, CTZ, ERYN, TTC | R6 | - | 1 | - | - | 1 | |
| Sub-total | - | - | 3 | 2 | 1 | 6 | |
| Private | ERY, KAN, TTC | R3 | - | - | 1 | - | 1 |
| Slaughter | AMP, ERY, TTC | R3 | - | - | 1 | - | 1 |
| House | AMP, CIP, ERY, TTC | R4 | - | 1 | - | - | 1 |
| Sub-total | - | - | 1 | 2 | - | 3 | |
| Butcher | ERY, KAN, TTC | R3 | 1 | - | - | - | 1 |
| House | AMP, ERY, SXT, TTC | R4 | - | - | 1 | - | 1 |
| AMP, CTZ, ERY, SXT, TTC | R5 | 1 | - | - | 1 | ||
| Sub-total | - | 2 | - | 1 | - | 3 | |
| Over all MDR (%) | - | - | 2 (14.3) | 4 (28.6) | 5 (35.7) | 1 (7.1) | 12 (85.7) |
Note: Ampicillin (AMP), Azithromycin (AZM), Cefotaxime (CTX), Ceftazidime (CTZ), Chloramphenicol (C), Ciprofloxacin (CIP), Colistin sulphate (CT), Erythromycin (ERY), Gentamicin (GEN), Kanamycin (KAN), Nalidixic Acid (NA), Sulfamethoxazole (SXT) and Tetracycline (TTC ), R3-R6, resistance to three, four, five, and six classes of antibiotics
Table 4: Multi-Drug Resistance (MDR) profile of E. coli O157:H7.
Foodborne infections related to contaminated foods of animal origin are major health concerns in developing countries including Ethiopia. Raw beef consumption is becoming an increasing trend in every corner of the country, mainly at butcher shops. Given that cattle are natural reservoirs of Shiga toxin-producing Escherichia coli strains, higher occurrence of E. coli O157:H7 is common in areas where fecal contamination continues such as lairage, slaughter hall, transportation trucks, carcass, and carcass in contact surfaces.
The overall prevalence of E. coli O157:H7 was 3.97% which is in agreement with the national prevalence estimate (4%) in Ethiopia. This finding was slightly higher than 2.4%, lower than 5.4%, and 8.6%.
Regarding to sample source, the prevalence of E. coli O157:H7 at the slaughterhouse level was 5.95% which is in line with the findings which is 5.7%. A lower prevalence than the present finding was reported from Ethiopia, the United Kingdom, and Ireland which reported 2.7%, 3.2%, and 3.0%, respectively. The variation in the prevalence could be attributed to the fact that there is a difference in slaughterhouse standards, hygienic practices of workers, sampling and isolation methodology, season, geographical origins, and the number of cattle.
In a previous study the occurrence of E. coli O157:H7 in butcher shops was observed to be higher, which was 13.3% (17/ 128) collected from butcher shops in central Ethiopia. Likewise, the prevalence was recorded in Ambo (19.1%) and Bishoftu (6.3%) butcher shops. The variation in the prevalence of E. coli O157:H7 in butcher shops could be due to the status of hygiene and sanitation practices of the butcher shops, sample size, and sampling technique. For example, positive isolates were recovered from the knife, (1/30), cutting board (3/30), and protective clothes (1/30). However, pooled sampling method, which was used in this study, recovers only one (1/92), showing sampling methods can be a big factor.
The occurrence of E. coli O157:H7 was significantly higher at the municipal slaughterhouse (8.3%) than at the private slaughterhouse (3.5%). The prevalence of E. coli O157 in the beef carcass was 3.3%, which is comparable to a report from Haramaya University slaughterhouse (2.65%, 3/113). The occurrence of E. coli O157 was significantly higher at the municipal slaughterhouse (8.3%) than at the private slaughterhouse (3.5%). The slaughterhouse facility, origin of the cattle, worker’s hygiene, and transportation of cattle from the origin to the slaughterhouse might have contributed to the differences in the occurrence of the pathogen. The slaughterhouse facility, origin of the cattle, worker’s hygiene, and transportation of cattle from the origin to the slaughterhouse might have contributed to this difference. A survey in Ethiopia discussed that public slaughterhouses have poor management and facilities than private abattoirs.
Regarding sample type, carcass and carcass contact surface swabs (12.5%) had a higher proportion of E. coli O157:H7 followed by fecal, content (3.3%) and beef samples (3.3%). The occurrence of E. coli O157:H7 in fecal, swab, and beef samples were also reported in different studies. In this study, even though the prevalence of E. coli O157:H7 was low, it was observed that there was no declining profile of prevalence from fecal (3.3%), carcass and carcass in contact surface with wastewater (12.5%) and beef sample (3.3%), demonstrating that cross-contaminations of carcass occur during slaughtering process which in overall reflect the general unhygienic conditions in employees, utensils and environmental sanitation of the slaughterhouse under study. It also indicates that the sanitary and hygienic measures at the slaughterhouses were ineffective against E. coli O157:H7.
Similarly to what is reported by which detected (3.6%, 4/110) E. coli O157:H7 on the surface of wooden cutting boards, this survey also isolated (1.1%, 1/92) E. coli O157:H7 from a pooled swab sample of butcher’s hand, knife and cutting board. However, in this study E. coli O157:H7 was not detected at the same time from the beef cut and beef contact surface samples swabbed from the same butcher shop.
In the present study, E. coli O157:H7 was isolated from slaughterhouse wastewater (1/12, 8.3%), which is alarming. This finding was lower than a previous study conducted in other countries. Reported a relatively higher 11% prevalence of slaughterhouses in Kayseri, Turkey. A similar higher prevalence of 16%, in Nigeria and 20.8% in Turkey was also reported. This finding notes the importance of ensuring the establishment of water treatment facilities for slaughterhouse wastewater efflux.
In general, the result of the present study was lower compared to previous studies conducted in Ethiopia and other countries. Considering the isolation methods employed, demonstrated the use of Enrichment Culture (EC) in modified buffered peptone water followed by Immunomagnetic Separation (IMS) for the isolation of E. coli O157:H7 from bovine faeces. Accordingly, of 1024 bovine rectal swabs, E. coli O157 was isolated from 23 by direct culture, and 84 by IMS including the 23 that were isolated by direct culture. Likewise, other researchers have also reported that IMS resulted in a greater detection rate for E. coli O157 in beef and bovine feces compared to enrichment plating. Thus, the absence of Immunomagnetic Separation (IMS) in the isolation procedure could be one reason for the low overall prevalence of this pathogen in the present study.
The Recto-Anal Junction (RAJ) of cattle is the principal site of colonization for E. coli O157:H7 and it was argued that E. coli O157:H7 was detected in a higher proportion from the intestinal mucosa proximal to RAJ than in the feces. Therefore, in the present study, the low prevalence of E. coli O157:H7 in the fecal sample observed may be associated with a low shedding profile of the pathogen by beef cattle included in this study.
The present study was also supported by other studies for the absence of the association between occurrences of E. coli O157:H7 and sample type. In contrast, other researchers have reported that sample type and E. coli O157:H7 have a significant association. This study has also considered an observational survey to assess the hygiene and sanitary practices of slaughterhouses and butcher shops and the overall slaughtering operations. Hygienic practices during beef production, processing, and distribution are essential to formulate preventive measures to mitigate the contribution of meat to foodborne diseases.
Unfortunately, this study was not able to collect samples from the truck, lairage, and hide. However, upon visual observation, the lairage was dirty and there were no compartments which increase fecal contamination of the hide due to close contact with animals. Previous works have revealed that the major source of beef carcass contamination was the hide of cattle entering the processing facilities. In a study done, a significant association was found between carcass swabs and skin swabs. Support these findings by demonstrating that antimicrobial interventions targeting cattle hide lead to drastic reductions in the rates of carcass contamination with E. coli O157:H7.
Stressful stunning and dragging of cattle on the floor, lack of clear demarcation between dirty and clean areas (municipal slaughterhouse), careless fisting and evisceration, sharing of knife, axe, and rasps, uncovered drainage line, lack of sequential decontamination at various stages, infrequent postmortem examination (municipal slaughterhouse), lack of hot water baths for hand washing and dipping of knives, uncleaned protective cloths (except in few butcher shops), infrequent hand washing were bad practices observed at the slaughterhouses and butcher shops. At slaughterhouses, workers transported the carcass from the hook bar to the vehicle on their shoulders. They used plastics gown which covered their back. However, the hygienic status of these protective clothing was not up to the standard required for abattoir workers. Personnel working at the slaughterhouses did not wear clean aprons, boots, and hair caps during meat processing. This might be the reason for the occurrence of E. coli O157:H7 in the beef sold at butcher shops. Hence, slaughterhouse workers should wear clean protective clothes, and also ensure their hands are always clean so as to produce high-quality beef. Microbial contamination of carcasses most likely occurred during evisceration. However, in the slaughterhouses under study, evisceration was conducted without considering the spillage of fecal material into the carcass from the gut.
With regard to hygiene and sanitation, it is a documented fact that a lack of education and training on food safety can contribute to unhygienic handling, processing, and display of meat at slaughtering places and at butcher shops. The slaughterhouses and butcher shops were not equipped with the necessary equipment which might enable them to maintain the general hygienic practice. For instance, there was no hot water for hand washing and knife dipping, clean towels, foot bath and separate rooms to process rumen and intestine (municipal slaughterhouse). Above all, an inadequate supply of tap water was one of the greatest challenges to maintain hygiene.
Thus, the relatively higher prevalence of E. coli O157:H7 on carcass swabs and beef in the current study might be due to the contamination of carcass with fecal material during the slaughter operation or from different contaminated materials and hands of meat handlers. Hence, because of an increasing trend of raw meat consumption throughout the country, this is an alarming situation. Studies clearly documented that; raw beef can harbor Shiga Toxin-producing E. coli (STEC) that causes diarrhea, hemorrhagic colitis, and Hemolytic Uremic Syndrome (HUS) in humans.
Antimicrobial resistance has come recently a challenge for ‘One Health’ due to the rapid emergence and spread of resistant bacteria among animals, humans, and the environment. Antimicrobial resistance may be developed spontaneously either by selective pressure or due to misuse by humans or overuse in feeding or treatment of beef cattle by owners. Resistance development may also associate with the exchange of resistance factors between related bacteria.
The present study indicated that all E. coli O157:H7 isolates were susceptible to three antimicrobials namely azithromycin, cefotaxime, and chloramphenicol which was in agreement with a local Ethiopian study isolates from goats. Likewise, ceftazidime, ciprofloxacin, gentamycin, kanamycin, nalidixic acid, and sulfamethoxazole showed greater activity against the isolates. In contrast, all E. coli O157:H7 isolates were resistant to tetracycline which again comes in parallel with the previous result. This is not surprising since tetracycline is often used as a first-choice antimicrobial for disease prevention and treatment in food animals and its widespread use has likely contributed to high rates of resistance. Closely related tetracycline resistance to this finding was also reported, from raw milk, in Ethiopia. Higher resistance to Erythromycin (92.8%) and Ampicillin (64.3%) was also observed among E. coli O157:H7 isolates. The current finding for tetracycline resistance disagrees with another study on E. coli O157:H7 isolated from feces and carcasses of cattle slaughtered in the abattoir.
Multiple antimicrobial resistance is becoming a common phenomenon among E. coli O157:H7 isolates. It may arise from the spread of genetic materials such as plasmids, integrons, and transposons drawn from different sources. In this study, multidrug resistance to three or more classes of antimicrobial classes was detected in 12 (85.7%) of the isolates. The most frequently observed resistance profile in the isolates was resistance to tetracycline in combination with erythromycin and ampicillin. Among the E. coli O157:H7 isolates, 42.8%, 28.6%, 7.1%, and 7.1% developed resistance to three, four, five, and six antimicrobials classes respectively.
A similar finding of multiple antimicrobial resistance on STEC strains has been documented in Ethiopia, and another part of the world. Statistically significant association was observed between the sample source (municipal slaughterhouse), and multiple antimicrobial resistance. Accordingly, isolates from municipal slaughterhouses were 5.84 times more likely to get multiple antimicrobial resistances than isolates from butcher shops. However, no statistical significance was established between sample type and multiple antimicrobial resistances which was in contrast with a previous report. Based on sample type, from fecal 28.6% (4/14), carcass and carcass contact swabs 35.7% (5/14), beef 14.3% (2/14), and from water and wastewater, 7.1% (1/14) multiple antimicrobial resistance profile was observed. The higher multiple antimicrobial resistance seen in this study might be due to the presence of only a few isolates tested for susceptibility compared to the overall study sample, the difference in sample source, variability of the resistant gene within isolates, and the type of antimicrobials used in the study. This result, multi-drug resistant wastewater isolates particularly, suggests slaughterhouse efflux would become a source of resistance pathogen to the environment.
E. coli O157:H7 was detected from the feces, carcass swab, and environmental samples with a slightly higher occurrence in carcass swab, possibly suggesting the role of hiding and gastrointestinal content as key sources of microbial contamination of the beef. The organism was also isolated from the butcher’s hand, knife, and cutting board (pooled sample) indicating a high potential for cross-contamination of the carcass during the slaughter operations, carcass transportation, loading, and unloading. Thus, control measures to reduce carcass contaminations were not absolute even if the prevalence is lower in this study. Isolation of E. coli O157:H7 from raw beef, which is resistant to multiple clinically important antimicrobials, highlights the potential threat to public health. This study has also attempted to assess the hygienic and sanitary practices in the slaughterhouses and butcher shops, and their respective workers. The results reflect that there were poor personal and general hygiene measures in place and that the workers did not focus on hygienic practices. A significantly higher occurrence of E. coli O157:H7 in municipal than in private slaughterhouses indicates less effective intervention measures for carcass contaminations in the former than the latter. The occurrence of multidrug-resistant E. coli O157:H7 isolate in slaughterhouse wastewater is a serious matter of concern as resistance genes are easily transferable into the environment as well as the food chain and human populations. Therefore, the consumption of raw beef may be an important source of E. coli O157 infections in the country and standard hygienic practices should be implemented in meat establishments.
Ethical Consideration
Verbal consent was obtained from the abattoir and retail meat shop owners before the commencement of sample collection. Moreover, ethical clearance was obtained from the animal research ethical and review committee of the college of veterinary medicine and agriculture of Addis Ababa university (Ref. No: VM/ERC/14/02/14/2022).
The authors are grateful to Addis Ababa university for its financial support. The authors would like also to thank the abattoir and retail meat shop owners for their support during sample collection.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Abunna F, Yimana M, Waketole H, Beyene T, Megersa B (2025) Detection and Antimicrobial Resistance Profile of E. coli O157:H7 from Slaughterhouses and Butcher Shops in Bishoftu Town, Central Oromia, Ethiopia. Food Microbial Saf Hyg.10:328.
Received: 22-Dec-2022, Manuscript No. JFMSH-23-21121; Editor assigned: 27-Dec-2022, Pre QC No. JFMSH-23-21121(PQ); Reviewed: 10-Jan-2023, QC No. JFMSH-23-21121; Revised: 13-Mar-2023, Manuscript No. JFMSH-23-21121; Published: 10-Feb-2025 , DOI: 10.35248/2476-2059.25.10.328
Copyright: © 2025 Abunna F, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.