Transcriptomics: Open Access
Open Access
ISSN: 2329-8936
ISSN: 2329-8936
Research Article - (2023)Volume 9, Issue 1
Objective: Given that endurance exercise can have a huge impact on nonelite athletes, this study set out to analyze differentially expressed genes and pathways before and after a marathon, then subsequently assess which body systems may be deregulated during such activity.
Methods: The study included 60 nonelite athletes (42 men and 18 women) participating in the Barcelona Marathon. Blood samples were extracted at three different time points: before the marathon at baseline levels (START), immediately upon completion (FINISH), and 24 hours after its completion (24REST). Differential gene expression, GO term, and KEGG pathway analyses were conducted on the samples from each of the groups and three different comparisons made: C1 (START vs. FINISH), C2 (FINISH vs. 24REST), and C3 (START vs. 24REST).
Results: The values for differential gene expression, GO terms, and KEGG pathways, respectively, were 9534, 162, and 61 in START vs. FINISH; 9454, 131, and 59 in FINISH vs. 24REST; 454, 14, and 8 in START vs. 24REST. When expression immediately after the marathon (FINISH) was compared with the other two groups (C1 and C2), we observed significant enrichment of terms related to the immune system, mitochondria, inflammatory markers, viral transcription and replication, reactive oxygen species, and lipid metabolism. Furthermore, upon comparing pre-marathon expression with levels 24 hours after its completion, the enriched GO terms were associated with mitochondrial activity, reactive oxygen species, and lipid metabolism.
Conclusion: Performing strenuous exercise deregulated immune system function, inflammatory markers, and mitochondrial terms, introducing a higher risk of infection in the period after the marathon, and it could alter the oxidation environment and lipid metabolism. While gene expression did not fully recovery 24 hours after the race, it was significantly closer to the baseline values than it was immediately after exercising.
Endurance exercise; Gene expression; RNA-seq; Next generation sequencing
Previous studies in sports science suggest that regular exercise is beneficial for our health, for example, it reduces the chances of developing heart disease [1-4]. Nevertheless, the consequences of performing exhausting endurance activities remain unclear. For example, a study looking at Tour de France participants found a significantly lower rate of cardiovascular events and longer life expectancy compared to age-matched, ‘ordinary’ French men [5]. However, pushing one’s body to the limit and constantly subjecting it to intense training, such as running a marathon, can have the opposite effect [6,7]. A possible explanation for these apparently contrasting situations is that other covariates may trigger our bodies to adapt to intense exercise. One hypothesis suggested that these differences may be due to age and training conditions [8].
To better understand what happens in the human body, the present study analyzed changes in the gene expression of subjects after they had run a marathon. Previous work focusing on gene expression during an ultramarathon reported a significant impact on the immune and inflammatory systems [9]. Furthermore, differentially expressed pathways were linked to protein synthesis repression, an altered immune system, and infectious disease-related mechanisms. Some studies suggest that strenuous exercise can induce immune system dysfunction and increase the risk of infection [10,11], while endurance activities could also have a direct impact on fatty acid metabolism. High-intensity training may increase the prevalence and severity of coronary atherosclerosis [12] and modify insulin resistance in humans [13].
Consequently, research into the impact of intense sports on health, both at a professional or amateur level, has grown recently, with a particular focus on the changes that this practice may cause in our genome so that we may mitigate any negative effects. Therefore, the aim of this study was to assess the effect of an abnormally intense physical effort on gene expression in blood samples. This was done by identifying genes with significantly different expression levels after the subjects had run a marathon; then studying the genes’ biological implications through pathway and gene ontology term analyses and assessing their significance during the race; and finally defining their expression recovery levels 24 hours after the strenuous exercise.
Sample collection
We extracted 2.5 mL of whole blood from 60 nonelite athletes (42 men and 18 women) before and after they participated in the 2016 Barcelona Marathon (42 km running). The samples were collected immediately before the start of the race (START), immediately after the race (FINISH), and 24 hours after completing the marathon (24REST).
The subjects were aged between 20 and 55 years; their heights and weights were also recorded. Subjects trained an average of 7.5 hours/week. All the steps in the workflow are summarized in Figure 1.
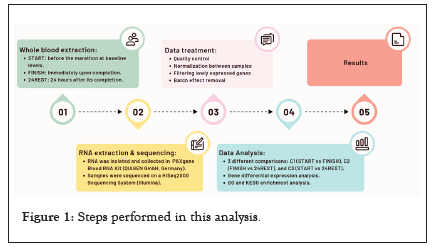
Figure 1: Steps performed in this analysis.
RNA extraction
Intracellular RNA was isolated from each subject’s whole blood sample and collected in PAXgene Blood RNA Tubes following the instructions of the PAXgene Blood RNA Kit (QIAGEN GmbH, Germany). Total RNA was stored at -80 °C until used. The FINISH time point samples were collected in a pavilion close to the marathon’s finish line that was perfectly equipped to conduct blood extractions. The START and 24REST samples, on the other hand, were obtained at the Hospital de la Sant Creu i Sant Pau. The samples were sent to Banc de Sang I Teixits (BST), Barcelona’s blood and tissue bank, for RNA extraction and sequencing. RNA samples were prepared for sequencing with the Illumina TruSeq sample preparation kit (Illumina, San Diego, CA) and using a Globin Block pack according to the manufacturer’s instructions and sequenced on a HiSeq2000 Sequencing System (Illumina).
Prior (to check either RNA integrity or RNA quality) and posterior (to ensure samples accomplish several parameters, Table S1) quality control checks were performed to discard samples that lacked sufficient quality for the following analysis.
Gene count
The 74-bp sequenced, paired-end reads were mapped to the GRCh38 reference genome using the STAR algorithm as the aligner and the standard parameters of the pipeline and salmon for gene quantification. Genes were annotated in accordance with Ensembl annotation.
Subjects for whom a valid count matrix could not be retrieved from Nextflow at any of the three time points (either due to difficulties collecting blood samples or extracting RNA or because of the low quality of the sequencing output) were discarded in this step.
The following analyses were all performed with R. Packages were used to filter genes with low expression levels and implement sample normalization [14,15]. Normalization was done by calculating scaling factors to convert raw library sizes into effective library sizes considering the library length. Each sample was corrected with the normalization factor after calculating the counts and the correction was included in the analysis. Other authors [16] have also discarded genes with low expression levels from the analysis to increase confidence and the number of Differently Expressed Genes (DEGs) observed. In this study, we used a function from the edgeR package, only keeping those genes with at least minimum count reads in all samples.
As not all samples were sequenced in the same run, we considered every group of samples sequenced together as a single batch. Sva, and more specifically its combat function, was used to correct any noise or batch effect that this method might introduce.
Fitted linear regression models of gene expression
The filtered, normalized matrix of counts described above was used as input for the downward analysis.
First, we performed a Principal Component Analysis (PCA) and then used the visualization of the individuals’ coordinates to consider covariates or clusters of data in the following steps.
The analysis was conducted using the Limma [17] package to build linear models for RNA-Seq data. The design matrix was built considering the covariates found with the abovementioned method:
Design matrix=0+Vg+Vs+Vi (1)
Where 0 corresponds to the matrix without an intercept, Vg indicates a group (START, FINISH, 24REST), and Vs. and Vi refer to the sex and individual variables, respectively.
The matrix was built without an intercept because we did not intend to compare the expression of two of the groups against a control, but rather to compare all three time points with the others: START vs. FINISH (FINISH levels –START levels); FINISH vs. 24REST (FINISH levels-24REST levels); START vs. 24REST (24REST levels - START levels).
Differential Expression Gene (DEG) analysis
After creating the linear model, we applied the lmFit function from sva package to fit a linear model to each gene. Given these linear models, eBayes was used to compute moderated t-statistics, moderated F-statistic, and log-odds of differential expression by empirical Bayes moderation of the standard errors toward a common value. This method was used to measure any quantitative changes in expression levels between the experimental groups.
All p-values were corrected by applying the Bonferroni multiple comparison correction method [18]. Bonferroni is a very conservative method that reduces the false positive rate and was performed in all the analyses carried out in this study.
The following tests were applied three times, once to each comparison of the expression values at different time points. For convenience, these comparisons will be referred to as C1 (START vs. FINISH), C2 (FINISH vs. 24REST), and C3 (START vs. 24REST).
Gene ontology analysis
A Gene Ontology (GO) analysis was performed using the GOstats package [19] to test for both over- and underrepresented GO terms. The significant genes derived from the linear models mentioned in the previous section (e.g., genes with a corrected p-value <0.05) were used as the input in the GO analysis for each of the compared groups (C1, C2, and C3). A conditional hypergeometric test was used to identify relationships among the GO terms. This package, given a subclass of HyperGParams, computed hypergeometric p-values for over- or underrepresentation of each term in the given category (gene ontologies in this case) among the specified gene set.
The results from the GOstats package were filtered according to three main parameters: the total number of genes included in the GO term (size), the number of observed genes from the input population (count), and the probability of finding all those genes just by chance (odds ratio) and were represented using GOplot package [20].
KEGG pathway analysis
We used the same methodology as described in the previous section and the hypergeometric test was conducted using the signature Search package [21]. More specifically, we applied the enrichKEGG2 function, which returns KEGG pathway enrichment results when given a vector of gene identifiers.
After the checking the quality and filtering the samples, all analyses were performed on 60 (42 men and 18 women) of the initial population of 78 participants. Moreover, after taking out the genes with low expression levels, the initial input matrix contained 180 samples (3 time points for 60 subjects) and 14,235 genes (with Ensembl annotation).
We generated a heatmap using the gplots package [22] for the top 500 most variable genes in Figure 2, which included hierarchical clustering with a complete linkage method.
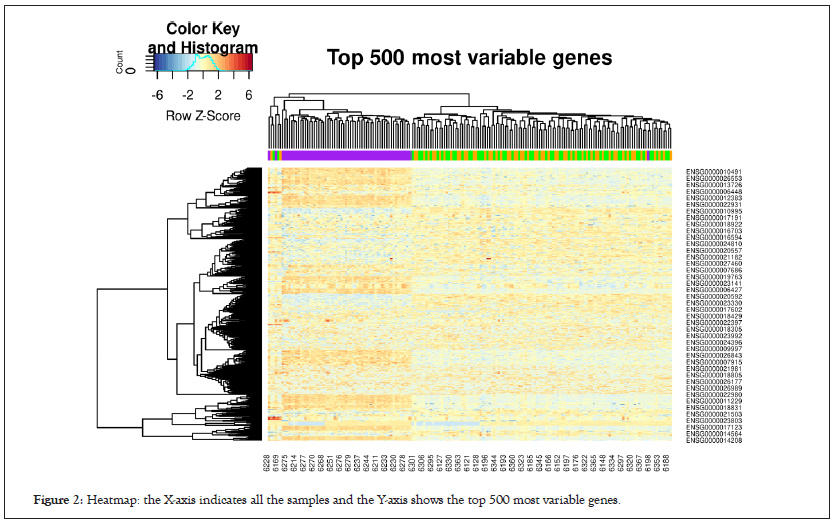
Figure 2: Heatmap: the X-axis indicates all the samples and the Y-axis shows the top 500 most variable genes.
The initial results suggest a clear difference between the FINISH samples (purple) and the STAR and 24REST samples (green and yellow).
Identifying covariates
If we look at dim2 (X-axis), there was a clear separation between the START and 24REST samples compared to the FINISH samples. With respect to the covariates, sex was identified as a parameter of inter-variability given the clear difference in dim1 between females and males. Moreover, intra-variability between samples also needs to be considered (Figure 3).
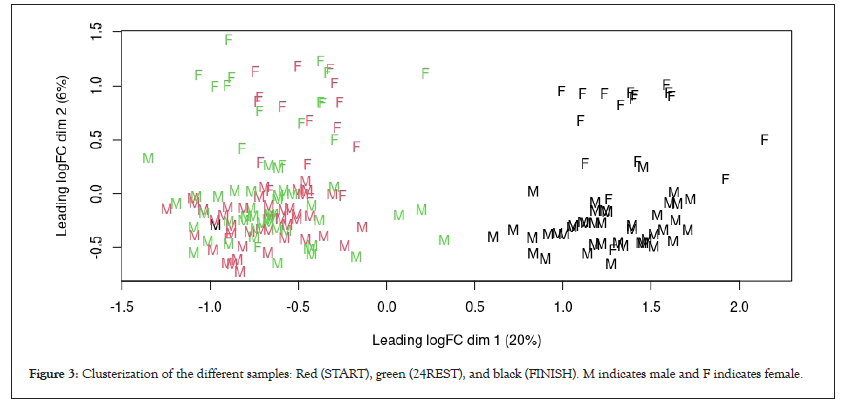
Figure 3: Clusterization of the different samples: Red (START), green (24REST), and black (FINISH). M indicates male and F indicates female.
DEGS
Differential gene expression analysis revealed that 9534 of the initial 14,144 genes were Differentially Expressed (DE) in C1, 9454 in C2, and only 454 in C3. Given how the groups were compared, the overexpression in groups C1 and C2 suggests that expression was significantly higher after the race (i.e., after strenuous exercise) than when at rest. While the overexpression of gene levels in comparison C3 indicates there was significantly higher level of gene expression 24 hours after the race compared to baseline levels (which suggests that subjects still had not recovered baseline expression levels after 24 hours rest).
We also performed a deeper analysis of the DE genes for each group comparison, with a particular emphasis on the biological function of the most significant genes.
START vs. FINISH (C1)
The genes in this comparison with significantly different expression levels were mainly associated with immune cell markers, chemokines, and interleukins (Table 1). Of the immune cell markers, CD48, CD19, LRRC8B, LRRC8C, and LRRC8D genes were found to be downregulated. The first two genes are involved in the B cell life cycle; in fact they are reliable markers of B lymphocyte activation and pre-B cells, respectively. The other three genes also play a fundamental role in B cell maturation and belong to the T cell activation leucine repeat-rich protein family. CD16 and CLEC10A, on the other hand, were significantly overexpressed. CD16 is a surface antigen preferentially expressed in monocytes, while CLEC10A is involved in the body’s inflammatory and immune response. CLEC10A mRNA expression is observed in intermediate monocytes, but most of the expression occurs in dendritic cells [23].
| ID | logFC | Adj. p-value |
|---|---|---|
| LRRC8B | -1687 | *** |
| CCR9 | -1,12 | *** |
| IL4R | -1225 | *** |
| IL10RA | -1091 | *** |
| SOCS4 | 1621 | *** |
| NADSYN1 | 0,6154 | *** |
| CD48 | -643 | *** |
| IL32 | -0,8615 | *** |
| CCR1 | 1453 | *** |
| IL6ST | 1033 | *** |
| IL6R | 1501 | *** |
| IL10RB | 1668 | *** |
| IL15 | -1578 | *** |
| LRRC8D | -0,9206 | *** |
| CCR5 | -1123 | *** |
| SOCS6 | 0,6181 | *** |
| CCL5 | 744 | *** |
| SOCS5 | 0,5578 | *** |
| GPX1 | -0,5111 | *** |
| CCR10 | -0,5314 | *** |
| ICOS | 0,4613 | *** |
| CCR6 | -0,7067 | *** |
| SOCS2 | 0,3546 | *** |
| GPX3 | -0,9665 | *** |
| SOD2 | -0,4221 | *** |
| CCR4 | -0,8518 | *** |
| CD19 | -246 | *** |
| NADK | -0,4668 | *** |
| LRRC8C | -0,4983 | *** |
| CCR2 | 0,6099 | *** |
| DNMT3A | 0,6606 | *** |
| SOCS1 | 0,5241 | * |
| CLEC10A | 0,1289 | - |
Note: (***), (**), and (*) indicate adjusted p-values (Bonferroni) of <0.001, <0.01, and <0.05, respectively.
Table 1: Differentially expressed genes related to immune system (cell markers, cytokines, and IL), ROS (Reactive Oxygen Species) environment, and mitochondria organelle in the START vs. FINISH comparison (C1).
As for the chemokines, most of the downregulated genes were expressed by T cell lymphocytes (CCR4, CCR5, CCR6, CCR9, and CCR10), whereas some genes related to the family of suppressors of cytokine signaling proteins were overexpressed (SOCS1, SOCS2, SOCS4, SOCS5, and SOCS6). Conversely, not all the genes related to cytokines were downregulated. Some genes, such as CCL5, which exhibit chemotaxis for T cells, eosinophils, and basophils but also induce the proliferation and activation of certain Natural Killer (NK) cells [24], and important specific cytokine receptors for monocytes, such as CCR2 and CCR1, were found to be overexpressed.
The last family of genes that stood out as DEGs was Interleukins (IL). The top downregulated IL were IL-4, IL-32, and IL-15. IL- 15 exhibits a broad activity and induces the differentiation and proliferation of T, B, and natural killer cells [25]. IL-32 is a pro- inflammatory cytokine involved in autoimmune diseases, such as rheumatoid arthritis [26], but it can also help protect the host against certain respiratory diseases, including tuberculosis [27]. Finally, IL-4 is an anti-inflammatory cytokine that inhibits IL-6 synthesis. However, the IL that we found to be statistically overexpressed were IL-6, a cytokine produced by myocytes when muscles contract [28], and IL-10 related protein, although not all the IL-10 interleukins were overexpressed. In fact, while ICOS (a protein that superinduces IL-10 synthesis) and IL10RB showed significantly higher expression levels in the FINISH group than the START group, IL-10RA (an IL-10 receptor) was downregulated in the same conditions.
As additional information, supplementary Tables S2 and S3 contain the DE genes related to Th1 and Th2 cells, respectively. These cells play an important role in immunity and have also been found to be deregulated in similar studies [29].
Table 1 also includes certain proteins that regulate oxidative stress, including the glutathione peroxidase family (GPX1, GPX3) and iron/manganese Superoxide Dismutase (SOD2), which were also downregulated. DNMT3A, which is an important gene related to reducing the oxidative environment, however, was overexpressed.
As such, we also detected several of the DEG inflammatory markers described in other studies; 15 out of 23 inflammatory markers reported elsewhere [30] were significantly differentially expressed in the present study (Table 2).
| ID | logFC | adj. p-value |
|---|---|---|
| IL4R | -1,418 | *** |
| CXCL16 | 1,115 | *** |
| IL1B | -1,087 | *** |
| IL6ST | 1,138 | *** |
| IL6R | 1,498 | *** |
| IL1R1 | 0,6862 | *** |
| IL10RB | 1,477 | *** |
| CCL5 | 0,7068 | *** |
| TGFBRAP1 | -0,7439 | *** |
| IL1RN | -0,6001 | *** |
| TNF | -0,5945 | *** |
| TGFBR3 | 0,5307 | *** |
| CCR4 | -0,7112 | *** |
| HSPA6 | -0,6677 | *** |
| CCR2 | 0,5805 | *** |
| GATA3 | 0,1996 | - |
| GPX7 | -0,1831 | - |
| TLR2 | 0,09307 | - |
| IFNGR2 | 0,1658 | - |
| SOD1 | -0,1404 | - |
| TLR4 | 0,2753 | - |
| GPX4 | -0,0576 | - |
| MMP9 | -0,01823 | - |
Note: (***), (**), and (*) indicate adjusted p-values (Bonferroni) of <0.001, <0.01, and <0.05, respectively.
Table 2: Table with differentially expressed genes related to inflammatory markers in the START vs. FINISH comparison (C1).
FINISH vs. 24REST (C2)
The results of the C2 comparison were very similar to those of C1. All the genes reported in C1 were also differentially expressed in C2 (Tables S4-S6). However, it is noteworthy that one of the top DE genes in C1 (CLEC10A) was not significantly DE in C2, as shown in Table S7.
START vs. 24REST (C3)
As mentioned above, only 454 genes were deregulated in C3, which is less than 5% of the DE genes in the other two comparisons. Not only was the number of significant DE genes much lower, but the genes were also different from those found in the other two comparisons (Tables S8-S11).
The top 10 DE genes for C3 are summarized in Table 3. Some cell progression inhibitors, such as CDKN1A/P21 and PPP6C, were found to be overexpressed. Furthermore, CDK17, which is involved in transcription initiation and DNA repair, was also upregulated.
| ID | logFC | adj. p-value |
|---|---|---|
| NFIC | 0,3923 | *** |
| CDKN1A | 0,3886 | *** |
| MANEAL | -0,299 | *** |
| ZFAND5 | 0,3448 | *** |
| RSF1 | 0,2391 | *** |
| PPP6C | 0,2812 | *** |
| CDK7 | 0,3916 | *** |
| PFKM | -0,3182 | *** |
| RFX3 | 0,3702 | *** |
| EBNA1BP2 | 0,3166 | *** |
Note: (***), (**), and (*) indicate adjusted p-values (Bonferroni) of <0.001, <0.01, and <0.05, respectively.
Table 3: The top 10 differentially expressed genes in START vs. 24REST comparison (C3).
Nevertheless, not all the overexpressed genes were involved in cell life functions. Some genes related to viral expression, including NFIC (which is involved in cell transcription and acts as a replication factor for adenovirus DNA replication) and RFS1 (which facilitates the transcription of hepatitis B virus genes), were also overexpressed.
Furthermore, we found genes associated with protein synthesis and degradation. For example, ZFAND5 (which is involved in protein degradation via the ubiquitin–proteasome system, besides playing a role in protein degradation during muscle atrophy) and EBNA1BP2 (involved in processing the 27S pre-rRNA). Finally, RFX3, an inflammatory gene previously reported as being overexpressed during exercise [31], was found to be overregulated.
By contrast to those genes, two of the top DE genes in the C3 comparison were downregulated: MANEAL and PFKM. It is interesting to highlight that a relationship between PFKM and exercise intolerance has previously been reported in PFKM knockdown mice (Table 3) [32].
Gene ontology term enrichment analysis
After performing conditional GO analysis and filtering the results by size and counts, 162 enriched GO terms were found in C1, 131 in C2, and 14 in C3. A complete list of all GO terms is given in Table S12 for C1 and S7 for C2.
Both comparisons returned similar results, while the top results for FINISH vs. 24REST are shown in Figure 4a. The top enriched GO terms were related to mitochondrial functions, T and B cell activation (which were downregulated in the FINISH samples), and virus-related GO terms (which were upregulated, in this case) (Figures 4a-4c).
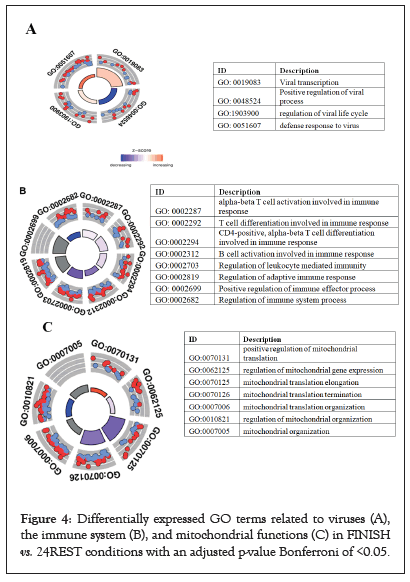
Figure 4: Differentially expressed GO terms related to viruses (A), the immune system (B), and mitochondrial functions (C) in FINISH vs. 24REST conditions with an adjusted p-value Bonferroni of <0.05.
The results of the comparison C3, START vs. 24REST, are shown in Table 4. Most of the enriched GO terms were related to energy generation processes, such as the electron transport chain or ATP synthesis.
| Terms | Adj.p-value |
|---|---|
| Chromatin-mediated maintenance of transcription | *** |
| Regulation of mitotic spindle assembly | *** |
| Mitochondrial respiratory chain complex I assembly | *** |
| Mitochondrial electron transport | *** |
| ATP synthesis coupled electron transport | *** |
| Translation initiation | *** |
| SRP-dependent cotranslational protein targeting to membrane | *** |
| Establishment of protein localization to endoplasmic reticulum | *** |
| Nuclear-transcribed mRNA catabolic process | *** |
| Electron transport chain | *** |
| Viral transcription | ** |
| Protein targeting to membrane | * |
| ATP metabolic process | * |
| Endoplasmic reticulum to Golgi vesicle-mediated transport | * |
Note: (***), (**), and (*) indicate adjusted p-values (Bonferroni) of <0.001, <0.01, and <0.05, respectively.
Table 4: Differentially expressed GO terms in START vs. 24REST comparison (C3).
KEGG pathway enrichment analysis
The KEGG pathway analysis was conducted as explained in the Materials and methods section, using the DE genes as input. Akin to the method for the GO term analysis, the pathway analysis was applied once at each of the time comparison points (C1, C2, and C3).
The total number of pathways enriched with DE genes were 61, 59, and 8 for comparisons C1, C2, and C3, respectively. Tables S13 and S14 show the total list of pathways for comparisons C1 and C2, while the list of enriched pathways for C3 is shown in Table 5.
| Description | Adj. p-value |
|---|---|
| Prion disease | *** |
| Thermogenesis | *** |
| Chemical carcinogenesis - reactive oxygen species | *** |
| Oxidative phosphorylation | *** |
| Parkinson’s disease | *** |
| Diabetic cardiomyopathy | *** |
| Nonalcoholic fatty liver disease | ** |
| Ribosome | ** |
Note: (***), (**), and (*) indicate adjusted p-values (Bonferroni) of <0.001, <0.01, and <0.05, respectively.
Table 5: Differentially expressed KEGG pathways in the START vs. 24REST comparison (C3).
The pathway analysis results closely resembled those of the GO term analysis, strengthening the significance of the findings. Moreover, the results of the C1 and C2 comparisons were also similar to each other but differed from C3. For comparisons C1 and C2, the pathways related to apoptosis, cellular senescence, mitophagy, and necroptosis were enriched (Tables S13 and S14). Further top enriched terms were a group of pathways related to lipid metabolism, including fatty acid metabolism, lipid and atherosclerosis, and some Sphingolipid signaling pathways.
In the FINISH vs. 24REST comparison (C2), insulin resistance and Vascular Endothelial Growth Factor (VEGF) signaling pathways, as well as those related to oxidative environment maintenance, were among the top significant results (Table 5).
Previous studies have investigated the effect of strenuous exercise on human gene expression. However, the present longitudinal study was carried out in nonelite athletes to measure the effects of an endurance event and determine if gene expression levels recovered after 24 hours. In general, the results suggest that strenuous exercise can induce an inflammatory response, activate an oxidative environment, and downregulate the immune system.
Previous studies have reported that endurance exercise induces an inflammatory environment, which indicates that long periods of strenuous exercise can generally lead to higher levels of inflammatory mediators and therefore may increase the risk of injury and chronic inflammation [33]. Our results mirrored these findings for 15 out of 23 inflammatory markers that were significantly differentially expressed between baseline (START) and FINISH levels (Table 2).
The immune system was also downregulated after performing the endurance exercise. The C1 (START vs. FINISH) and C2 (FINISH vs. 24REST) comparisons show that downregulated differentially expressed genes were enriched in GO terms and KEGG pathways related to B and T cells and other immune system components. This immune system downregulation may result in a higher risk of infection after performing endurance exercise, as reported by other authors [34,35]. What is more, infection and replication virusassociated terms were upregulated, which agrees with previous studies and indicates that endurance athletes experience extreme physiological stress during strenuous exercise, which is again associated with temporary immunodepression and a higher risk of infection, particularly Upper Respiratory Tract Infections (URTI) [36] given the immune system is weakened.
On the other hand, the C3 (START vs. 24REST) results reveal that 24 hours after performing endurance exercise, the immune system returned to almost the same levels as those before the race (baseline levels).
Despite everything the immune system undergoes, some monocyte markers, specific cytokines, and IL are overexpressed. Monocytes are activated during strenuous exercise, leading to acute inflammation and hypoxemia [37]. Furthermore, after performing long-exposure exercise, several genes that regulate oxidative stress, such as the glutathione peroxidase family (GPX1, GPX3) and iron/manganese Superoxide Dismutase (SOD2), were downregulated. This oxidative environment (ROS) could be the molecular link between monocyte chemotaxis and inflammatory pain [38]. Further studies are necessary to follow up on these observations and determine the impact and importance of monocytes in this type of exercise.
One gene that may play a more significant role in reducing the oxidative environment generated by strenuous exercise is DNMT3A, which was overexpressed in C1 (START vs. FINISH). DNMT3A expression in red oxidative muscle increases significantly following a bout of endurance exercise [39]. Muscle-specific Dnmt3a knockout mice have reduced tolerance to endurance exercise, accompanied by a reduction in oxidative capacity and mitochondrial respiration. Moreover, Dnmt3a-deficient muscle overproduces Reactive Oxygen Species (ROS), the main contributor to muscle dysfunction.
As mentioned in the Results section, apart from the inflammatory and immune systems, functions and genes related to mitochondria activity were among the top deregulated genes. One of the principal mitochondrial functions is to generate energy through the electron transport chain. Thus, we can infer that genes related to this organelle will be differentially expressed during endurance exercise because athletes need and consume more energy compared to baseline levels. Our results corroborated these expectations.
Another point of interest concerns comparison C3, that is, FINISH compared to 24REST samples, as baseline expression levels were not always fully recovered. Some of those enriched GO terms are associated with mitochondrial, electron chain, and ATP metabolism terms, which implies that the nonelite athletes did not have the same energy generation capacity 24 hours after the race as before starting.
Finally, some KEGG pathways related to diabetic cardiomyopathy and atherosclerosis were deregulated. Previous studies have reported a significantly higher rate of coronary artery calcification in long-term marathon athletes [40], and long-term male marathon runners may have paradoxically increased coronary artery plaque volume.
In conclusion, completing an endurance exercise, such as running a marathon, has a huge impact on gene expression levels and, consequently, the deregulation of several metabolic pathways and systems. With regard to immunity, downregulation of T and B lymphocytes lead to a temporary increase in the risk of infection after performing exhaustive exercise. On the other hand, monocytes may play a fundamental role during strenuous exercise, because their deregulation might be associated with lipid metabolism, inflammation, and the oxidative environment.
Nonelite athletes require such a huge amount of energy needed to run a race like this that their mitochondrial activity does not fully recover after 24 hours of rest. Some pathways related to atherosclerosis were also deregulated after finishing the race, so maybe protective measures should be taken before such an exhaustive effort.
This study may help establish future training routines or nutritional guidelines based on individual gene expression levels. What is more, the time to recover after strenuous exercise could be reduced through correct training, healthy routines, and specific gene expression information.
This paper and the research behind it would not have been possible without the exceptional institutional support of Exheus, Hospital Sant Pau de Barcelona, and Universitat Politècnica de Catalunya (UPC).
The study was conducted according to the Declaration of Helsinki. The ethical committee at Hospital de la Santa Creu i Sant Pau (Barcelona) approved the study. All participants provided informed consent. All data included in this study were managed under the umbrella of the General Data Protection Regulation (GDPR), to protect participants concerning the processing of personal data and the free movement of such data.
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
Citation: Condeminas PE, Antiga LG, Ros JB, Cardenas A, Sibila O, Perera-LLuna A, et al. (2023) Transcriptome Analysis in Response to Endurance Exercise in Non-Elite Marathon Runners. Transcriptomics: Open Access. 09:134.
Received: 20-Feb-2023, Manuscript No. TOA-23-21869; Editor assigned: 24-Feb-2023, Pre QC No. TOA-23-21869 (PQ); Reviewed: 10-Mar-2023, QC No. TOA-23-21869; Revised: 17-Mar-2023, Manuscript No. TOA-23-21869 (R); Published: 24-Mar-2023 , DOI: 10.35248/2329-8936.23.9.134
Copyright: © 2023 Condeminas PE, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.