
Journal of Clinical Chemistry and Laboratory Medicine
Open Access
ISSN: 2736-6588

ISSN: 2736-6588
Original Research Article - (2022)Volume 5, Issue 6
Objective: Poorly Differentiated Thyroid Carcinomas (PDTC) and Anaplastic Thyroid Carcinoma (ATC) are different subtypes of thyroid carcinoma characterized respectively by a medium to high degree of dedifferentiation. These rare tumor types account for the majority of deaths from thyroid cancer, having usually a poor prognosis and short life expectancy. Diagnosis is not always straightforward due to the high degree of dedifferentiation of these tumors. Affected patients, on the other side, need for quick and targeted clinical answers for an effective treatment. Recent studies utilizing next generation sequencing have brought light into the molecular landscape of these tumors, providing evidences to support a stepwise progression from differentiated thyroid carcinomas to anaplastic thyroid carcinomas.
Methods: We tested Illumina TruSight Oncology 500 comprehensive genome panel platform on a preliminary set of 10 PDTC and 8 ATC FFPE samples for the identification of molecular landscape and genetic pathways involved and dysregulated in the tumorigenesis of PDTC and ATC. The assay evaluates at the same time DNA and RNA specific alterations and allows the analysis of important biomarkers for the response to immune-checkpoint inhibitor therapy as tumor mutational burden and microsatellite instability. After the sequencing pipeline, results were evaluated and shared with Pierian Dx Genomic Landscape, for the generation of a complete clinical report.
Results: We were able to identify Tier I, II and III variants for all analyzed samples, biomarker values and fusions, picturing a cancer specific profile which might be useful for a clinician to stratify patient with complicated diagnosis history, in a relatively short turnaround time.
Conclusion: With one single comprehensive profiling approach, we were able to investigate these aggressive and poorly characterized thyroid cancer subtypes, in a more comprehensive and faster fashion, providing clinicians with a complete clinical report allowing them to discuss the clinic and therapeutic applications of the retrieved data for patient management.
Thymosin α1; TNBC; Treatment; Signaling pathway
Thyroid cancer is reported to be the most frequent endocrine malignancy, accounting for 3%-4% of cancers worldwide [1,2]. The incidence of thyroid cancer showed an increase in the last decades, with an estimated annual percentage change of 1.59 [3] with females accounting for most of the thyroid cancer burden (77, 22% of incidence) [3]. The development of more sensitive detection systems and diagnostic tools, as Ultrasound Scans (US) and other imaging techniques, together with an increased number of thyroidectomies and better histological and molecular sample examination [4] seems to be one of the reasons behind the rise in the newly diagnosed cases in the last 10 years [5]. This increased diagnostic rate contributes to the improvement of thyroid cancer characterization, allowing for a better understanding of tumorigenesis mechanisms and thyroid cancer progression and development. Thyroid Cancer (TC) includes a wide group of malignancies, developing from different tissue types. TCs are generally subdivided into two main TC subgroups: a large group of well differentiated neoplasms characterized by slow growth and high curability, and a small group of highly dedifferentiated tumors with poor prognosis [6]. The World Health organization classifies TCs in main 5 histological types: Papillary (PTC) and Follicular Thyroid Cancers (FTC), Poorly Differentiated (PDTC) and Anaplastic Thyroid Cancer (ATC), which originate from the follicular epithelial cells, producing thyroid hormone, while the fifth subtype, namely Medullary Thyroid Cancer (MTC), originates from parafollicular cells, responsible for the production of calcitocin [7-10]. TC types originating from follicular epithelial cells are the most common, with papillary carcinoma being responsible for 80% of diagnosed thyroid carcinoma cases, and follicular carcinoma for 10% [8,10]. PTCs and FTCs are grouped under the name of Differentiated Thyroid Cancers (DTCs), having usually a good prognosis and long-term survival rate. In fact, even though relapse rate is up to 30%, life expectancy of DTC affected patients is comparable to the general population one, after treatment [10,11], with a 5-year survival rate reaching 91.1% and 79.9% in PTC and FTC [7,12].
Although this good prognosis and good life expectancies, about 20% of DTCs patients are at high risk for complications and therapy failure because of tumor recurrence, failure to absorb radioiodine. In rare cases progression of well-differentiated tumors to poorly differentiated or anaplastic thyroid carcinomas might also be a reason for therapy failure and poor outcome of the patient [10]. These carcinomas are in fact aggressive tumor types associated with bad prognosis, high mortality rate (38% [13] and 100% respectively [14]), and an overall survival of 6 years for PDTCs and only 6 months for ATCs [12].
The mechanism of DTCs dedifferentiation is not completely characterized. Up to date two main models have been proposed to explain this transformation: a “stepwise transformation from DTC to ATC” and a “the novo model of independent transformation from PDTC and ATC”. This dedifferentiation process linking PDTCs and ATCs was investigated in a single patient based 2019 study of the Shangay Medical College [15]. Even though there are different evidences supporting ATCs arising de novo, the dedifferentiation process of ATC from pre-existing DTC is commonly accepted and it recognize in PDTCs an intermediate state in its progression [16,17].
Studies on PDTC and ATC tumorigenesis and biology are nowadays complex and suffer from the same pitfalls. These are in fact rare tumors and this makes it difficult to properly depict the biology and dedifferentiation mechanisms of these tumor types. Furthermore, clinical diagnosis and management of these tumors is not straightforward, since diagnostic criteria are not widely shared. In case of PDTCs, criteria of Turin consensus conference [18] or Memorial Sloan Kettering Cancer Center [19] are differentially used to make diagnosis in different centers. For what concerns ATC cases instead, there is a wide spectrum of histotypes making it challenging to perform a differential diagnosis with other cancers [12]. There is still a lack of effective therapy for PDTC and ATC affected patients, which represents an unmet clinical need that should be addressed. Poor prognosis and low survival rates after diagnosis, makes necessary to find rapid and complete diagnostic tool able to provide the clinician with valuable data for a better patient treatment and management.
Up to date, molecular targeted therapies are being tested and studied in human clinical trials [20-22], and clinicians are proposing genetically guided treatments for PDTC and ATC, led by new discoveries about their genetic landscape [12]. Different studies demonstrated in fact, that the molecular classification of TCs better explains its underlying characteristics than histological classification alone [23].
For all these reasons, the Synlab Molecular Biology unit together with University of Perugia and the Anatomical Pathology Unit of Synlab Italia decided to initiate a study for the identification of molecular landscape and genetic pathways involved and dysregulated in the tumorigenesis and progression of PDTC and ATC, to provide a comprehensive, rapid and informative clinical option for rare tumors with low survival rates.
We planned a preliminary study to characterize 10 PDTC and 8 ATC samples with a comprehensive genome profiling approach, working with Illumina TruSight Oncology 500 (TSO-500) analysis pipeline. TSO-500 pipeline consists of the parallel analysis of 523 genes related to oncogenesis and tumor progression, and 55 different fusion genes identifying known and unknown rearrangements. This comprehensive approach also evaluates two important biomarkers helping in selecting patient eligible for immune checkpoint inhibitor therapy: Tumor mutation burden [24] and Microsatellite Instability [25].
After the sequencing pipeline, results were evaluated and shared with Pierian Dx Genomic Landscape, for the generation of a complete clinical report in compliance with ASCO, AMP and ESMO guidelines.
Samples
The department of Medicine and Surgery of the University of Perugia provided us formalin fixed paraffin embedded tissues from 15 patients, comprising 10 PDTC patients and 5 ATC patients and the Anatomical Pathological unit of Synlab Italia supplied us 3 additional ATC FFPE samples to increase the study cohort and the statistical confidence of the acquired data.
Tumor diagnosis
Clinical data (gender, age, tumor size, lymph node and distant metastases and extrathyroidal infiltration) were collected analyzing patients’ charts and pathology reports. The diagnoses of all the thyroid lesions were reviewed and re-evaluated by the pathologists according to the World Health Organization’s criteria [8]. In detail, Turin criteria were adopted for the definition of PDTC [18]. The most representative paraffin block of each sample was selected for analysis. The study was conducted anonymously and in compliance with the principles of the Helsinki Declaration of 1975.
Nucleic acid extraction
DNA and RNA were extracted from the same biopsy using two different protocols, each one optimized for the extracted material. For DNA extraction, FFPE tissue sections underwent a phase of deparaffinization with xylene, followed by digestion with Buffer ATL and Proteinase solution at 56°C O/N. Digested DNA samples were than extracted via automated extraction method using QIAsymphony extraction technology with DNA mini kit (Qiagen, REF: 937236), following the Tissue LC 200 DSP protocol. Samples were finally eluted in 50 ul. Synlab Anatomical Pathology unit prepared 5-10 slides with samples tissue sections (5-6 micron thick) from which RNA was extracted after slides deparaffinization procedure with limonene. Nucleic acid extraction was performed manually using FFPET RNA Isolation Kit (Roche, REF: 06483852001) according to manufacturer’s protocol. RNA was eluted in a final volume of 30 ul.
Sample quantity and quality control
To ensure optimal assay performances, DNA and RNA samples extracted from FFPE specimens were quantitatively and qualitatively evaluated before undergoing library preparation. Qubit™ dsDNA BR Fluorimetric Assay Kit (ThermoFisher Scientific, REF: Q32850) was used for DNA quantification, performed following manufacturer’s protocol. Qubit™ RNA HS Assay Kit (ThermoFisher Scientific, REF: Q32852) was used for RNA quantification, following manufacturer’s protocol. A minimum input of 40 ng for DNA and RNA was used as input for library preparation. DNA sample quality was assessed using the Infinium FFPE QC Kit (Illumina, REF:15013664), according to manufacturer’s protocol. After Real-Time PCR assay, QC comparison between samples and positive control was evaluated, and only samples with ΔCq value ≤ 5 were considered qualitatively optimal. RNA samples quality check was performed evaluating RNA sample DV200 data after analysis at the Tape Station Analyzer D1000. High Sensitivity RNA ScreenTape System (Agilent, REF: 5067-5579, 5067-5580, 5067-5581) was used. Only samples with DV200 >20% (percentage of fragments above 200 bp size) were used for downstream applications.
DNA and RNA library preparation
Both DNA and RNA samples meeting quantitative and qualitative requirements underwent library preparation using the hybrid capture-based TSO-500 DNA/RNA Kit (24 Samples), plus PierianDx pipeline (Illumina, REF: 20040768). DNA library preparation begins with genomic DNA (gDNA) shearing using Covaris S220 technology (Covaris, REF: 500217) to obtain 200 bp DNA fragment. 12 μl of each diluted and purified gDNA sample were processed during the sonication. RNA library preparation begins instead with RNA retrotranscription using a two-steps retrotranscription process, performed according to Illumina protocol. Library preparation was performed following TSO-500 pipeline. Before beads-based normalization, samples were quantified using Qubit™ dsDNA HS Assay Kit (ThermoFisher Scientific, REF: Q32854) and qualitatively analyzed at the Tape Station Analyzer D1000 using High Sensitivity D1000 ScreenTape System (Agilent, REF: 5067-5584). 10 ul of every single DNA and cDNA library were mixed to prepare one DNA and one cDNA sequencing pool, and subsequently incubated for 2 minutes at 96°C for a denaturation step, blocked them via 5 minutes incubation on ice. DNA and cDNA pools were then mixed in a 4:1 ratio respectively. 10 ul of the denatured pool were transferred into a new microcentrifuge tube and 190 ul of HT1 buffer were added to it. After vortexing and centrifuging, 40 ul of diluted pool were transferred into a new tube and 1660 ul of HT1 buffer were added to dilute the sequencing pool (final libray concentration 1.5 pM). 2.5 ul of denatured 20 pM PhiX control was added. The sequencing run were performed on NextSeq 550Dx system (Illumina).
Bioinformatic analysis
The Illumina TSO-500 module comprises different settings helping the laboratory staff in setting up the run and performing the analysis. It supports run setup, sequencing, and bioinformatic analysis for the prepared DNA and RNA libraries [26]. The Local Analysis Software workflow on the NextSeq 550 Dx begins with the demultiplexing of BCL files and the software generates next the FASTQ files. These files are later used as input for the bioinformatic analysis performed with an Illumina application on BaseSpace Illumina server, namely TSO-500 Evaluation App. After the bioinformatic analysis is performed, according to manufacturer’s protocol, the DNA library analysis outputs include tumor mutational burden, variant call files for small and complex variants, microsatellite instability, and gene amplifications, while the RNA library analysis outputs include fusions and splice variant call files [26]. The TSO-500 DNA and TSO-500 RNA analysis workflow are quite complex and are schematically represented in Figures 1 and 2 respectively.
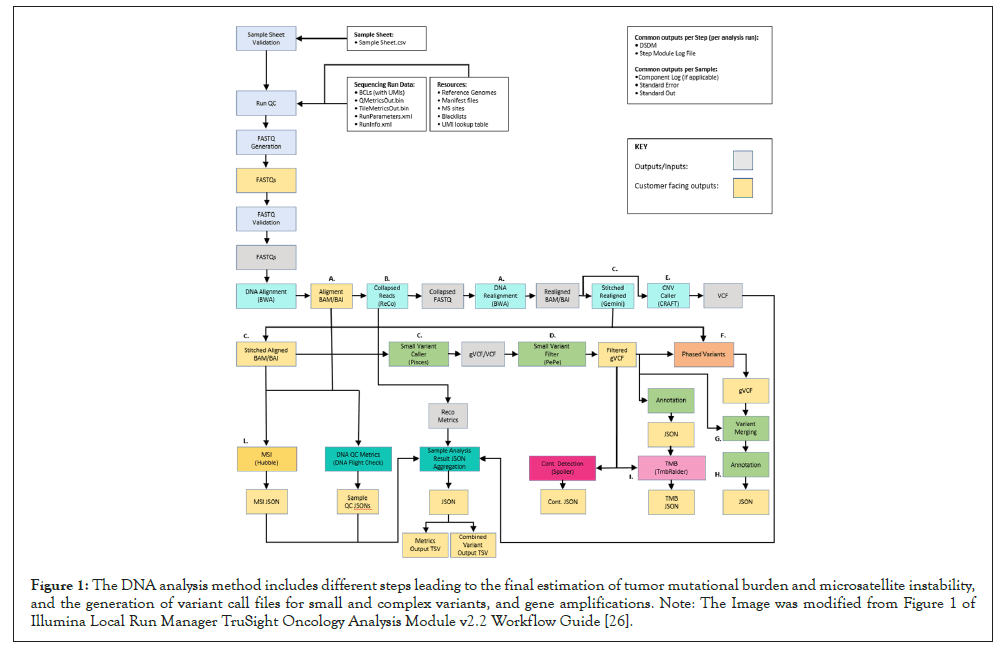
Figure 1: The DNA analysis method includes different steps leading to the final estimation of tumor mutational burden and microsatellite instability, and the generation of variant call files for small and complex variants, and gene amplifications. Note: The Image was modified from Figure 1 of Illumina Local Run Manager TruSight Oncology Analysis Module v2.2 Workflow Guide [26].
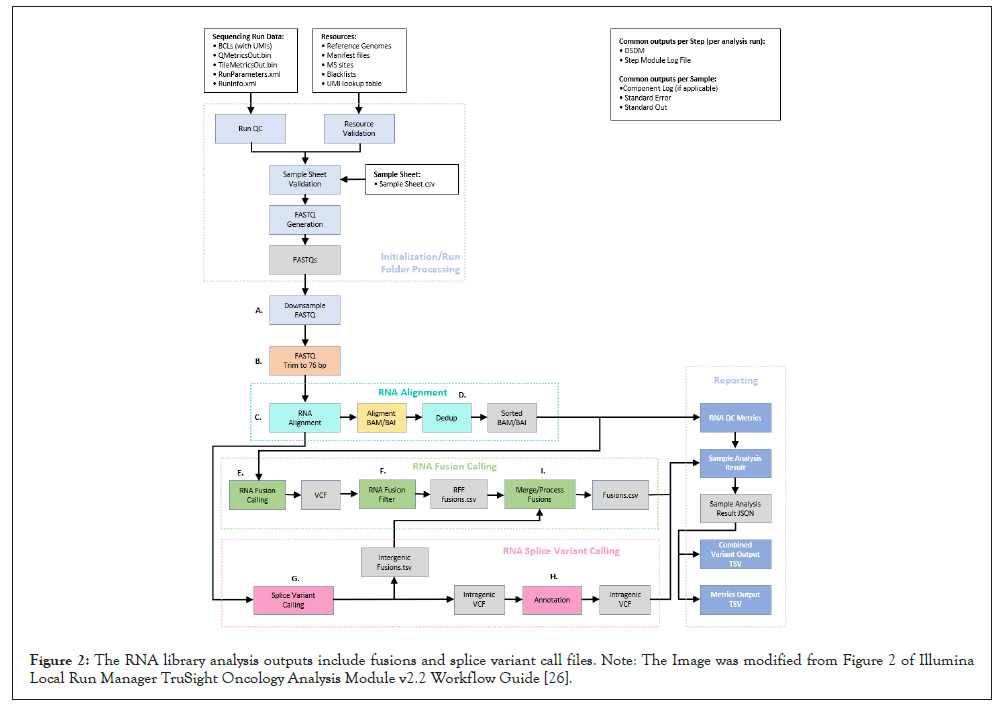
Figure 2: The RNA library analysis outputs include fusions and splice variant call files. Note: The Image was modified from Figure 2 of Illumina Local Run Manager TruSight Oncology Analysis Module v2.2 Workflow Guide [26].
Polymerase chain reaction analysis and Sanger sequencing for fusion confirmation
NTRK3-ETV6 fusion was confirmed by PCR amplification and Sanger sequencing. Primers for the amplifications were designed using Primer3 v.4.0 software, using as reference sequence, the contig sequence generated by the Illumina TSO-500 Evaluation App. Primer Forward (3’-CATTCTTCCACCCTGGAAAC-5’) was designed on ETV6 region proximal to the fusion breakpoint, and Reverse primer (3’-AGTCATGCCAATGACCACAG-5’) on the NTRK3 region proximal to the fusion breakpoint. Initial denaturation step is carried out at 95°C for 10 minutes and it’s followed by 40 cycles of denaturation-annealing-elongation according to these parameters: 95°C for 30 seconds, 57°C for 30 seconds and 72°C for 30 seconds. Final elongation step is carried out at 72°C for 7 minutes followed by 4°C in continuum. PCR was performed using AmpliTaq Gold™ DNA Polymerase (Applied Biosystems™, REF: 8080249). Sanger sequencing with PCR product was performed with a BigDyeTM Terminator Cycle Sequencing Kit (Applied Biosystems, REF: 4337455) using an ABI 3500XL Genetic Analyzer (Applied Biosystems).
Statistical analysis
GraphPad Prism software v5.0 (GraphPad) was used for all the statistical analysis. Two-tailed Mann-Whitney U-test was performed to compare total mutation rate (%) between PDTC and ATC independent groups subjected to the analysis. Two-tailed non-parametric t-test, not assuming data Gaussian distribution, was performed to compare TMB values between PDTC and ATC independent groups subjected to the analysis. The same statistical analysis was performed to compare MSI values between PDCT and ATC independent groups. Two-tailed Spearman non-parametric correlation test was used to test correlation between TMB and MSI values between ATC and PDTC samples separately. P values lower than 0.05 (P<0.05) were considered significant.
DNA and RNA quality parameters and metrics output
In this preliminary study, a total of 18 samples were analyzed, profiling both DNA and RNA for each sample. All samples derived from FFPE blocks. Sample minimum input was 40 ng for DNA and 40 ng for RNA and all samples analyzed met this requirement. Since extraction from FFPE tissues usually results in low quality and low yield material, samples suitability for the assay was tested both for extracted DNA and RNA. DNA sample quality was assessed using the Illumina Infinium FFPE QC Kit. Only samples with a Delta quantification cycle (Delta Cq) lower than 5 would pass the initial QC evaluation. All the available samples met these criteria, with a median Delta Cq value of 0.815. RNA samples quality was evaluated calculating sample DV200 value after analysis at the Tape Station Analyzer D1000. The DV200 parameter classifies degraded RNA by size helping in selecting samples suitable for the sequencing. Only samples with DV200>20% would pass the initial QC evaluation and be selected for the downstream analysis. All the available samples met these criteria, with a median DV200 value of 69, 39%. Final input and quality of the DNA processed samples are reported in Table 1.
| Sample name | DNA quantification (ng/ul) | Infinium FFPE QC (Delta Cq<5) | RNA quantification (ng/ul) | DV200 RNA QC (>20%) | Sample processability | DNA input (ng) | RNA input (ng) |
|---|---|---|---|---|---|---|---|
| PG2057-A4/17 | 12,9 | 0,63 | 1550 | 65,34 | ٧ | 100 | 100 |
| PG5315-A4/14 | 13,3 | 0,77 | 1370 | 63,41 | ٧ | 100 | 100 |
| PU5741-A9/17 | 21,2 | -0,27 | 335 | 41,84 | ٧ | 100 | 100 |
| PG28902-A3/18 | 21,7 | 0,57 | 825 | 78,43 | ٧ | 100 | 100 |
| PU4215-A1/12 | 39,9 | -1,06 | 398 | 61,55 | ٧ | 100 | 100 |
| PU7307-A3/13 | 11,1 | -0,14 | 760 | 58,94 | ٧ | 100 | 100 |
| PU7055-A2/14 | 7,37 | 0,86 | 197 | 69,39 | ٧ | 100 | 100 |
| PU14209-A4/17 | 3,61 | 0,76 | 160,5 | 77,73 | ٧ | 40 | 100 |
| 10-07-8383 | 9,85 | 2,94 | 182 | 73,15 | ٧ | 100 | 100 |
| 7143-a15/13 | 3,42 | 2,44 | 18,8 | 77,17 | ٧ | 40 | 100 |
| 11081-7/08 | 37,1 | 2,28 | 500 | 83,92 | ٧ | 100 | 100 |
| 15613-3/08 | 32 | 3,01 | 255 | 68,59 | ٧ | 100 | 100 |
| 20-I-59397 A5 | 32,2 | 0,94 | 475 | 75,25 | ٧ | 100 | 100 |
| 19-I-48528 A3 | 19,6 | 1,31 | 360 | 80,05 | ٧ | 100 | 100 |
| 19-I-48528 A10 | 28,5 | 2,29 | 425 | 84,69 | ٧ | 100 | 100 |
| PU 4940-A2/12 | 2,5 | 0,34 | 193 | 54,72 | ٧ | 60 | 100 |
| 430-2/10 | 2,18 | 1,60 | 174 | 55,72 | ٧ | 80 | 100 |
| PU1859-A1/15 | 1,64 | 0,35 | 360 | 34,84 | ٧ | 60 | 100 |
Table 1: Sample quantitative and qualitative control. The table shows whether the processed samples meet the required quality and quantity recommendations for proceeding with TSO-500 pipeline, and the DNA and RNA input used for the library preparation.
After the bioinformatics analysis pipeline of TSO-500 Local App, a metrics output file is generated. This report provides values to determine if run quality results meet the quality control criteria [27]. This file reports general sequencing run QC metrics, DNA library QC metrics and RNA library QC metrics.
The sequencing run QC metrics reports whether the run was successfully completed or whether errors occurred during the sequencing steps. In the 3 performed runs no errors occurred, and the sequencing was successfully completed, with a mean percentage of reads passing filter of 94.4% (PTC_PF_READS>80%). DNA sample QC metrics inputs are alignment data, read collapsed BAM, indel realignment, read stitching BAN and CRAFT normalized BinCount.tsv files. With these data the file generates a report evaluating: contamination score (≤ 3106) and contamination p-value (if contamination Score>3106, contamination p-value should be ≤ 0.049); the median exon coverage (≥ 150); the percent exon bases with 50X fragment coverage (≥ 90.0); the median insert size (≥ 70) and the usable MSI sites for MSI calling (≥ 40). These metrics were analyzed, and all the sequenced DNA samples met the quality metrics requirements, except for sample 19-I-48528 A10, which has not been considered for Single Nucleotide Variant (SNV), Tumor Mutation Burden (TMB) and Microsatellite Instability (MSI) evaluation. RNA sample QC metrics inputs are RNA alignment data. With these data the file generates a report evaluating: the median CV for all genes with median coverage >500X, which are more likely to be highly expressed (≤ 93), the median insert size (≥ 80) and the total on target reads mapping on the target region (≥ 9000000). These metrics were analyzed, and all the sequenced RNA samples met the quality metrics requirements. DNA samples QC metrics data are reported in Table 2, while RNA samples QC metrics data are reported in Table 3.
Control SNV, TMB and MSI calls reproducibility
| Samples | DNA library QC metrics | QC metrics for small variant calling and TMB | QC metrics for MSI | |||
|---|---|---|---|---|---|---|
| Contamination score | Contamination p-value | Median insert size (bp) | Median exon coverage (Count) | PCT exon 50X (%) | Usable MSI sites (Count) | |
| Lower Limits (LSL) Guidelines | 0 | 0 | ≥ 70 | ≥ 150 | ≥ 90.0 | ≥ 40 |
| Upper Limits (USL) Guidelines | ≤ 3106 | ≤ 0.049 | NA | NA | NA | NA |
| PG2057-A4/17 | 9859 | 0.001 | 105 | 477 | 99.5 | 119 |
| PG28902-A3/18 | 359 | 1 | 107 | 666 | 99.6 | 124 |
| PG5315-A4/14 | 473 | 1 | 98 | 445 | 99.5 | 115 |
| PU4215-A1/12 | 195 | 1 | 115 | 568 | 99.6 | 122 |
| PU5741-A9/17 | 200 | 1 | 120 | 761 | 99.5 | 123 |
| PU7055-A2/14 | 7548 | 0 | 91 | 392 | 99.5 | 116 |
| PU7307-A3/13 | 351 | 1 | 98 | 506 | 99.6 | 119 |
| 11081-7/08 | 9572 | 0.001 | 92 | 286 | 99.4 | 103 |
| 15613-3/08 | 859 | 1 | 84 | 209 | 99.1 | 90 |
| 20-I-59397 A5 | 145 | 1 | 121 | 738 | 99.6 | 119 |
| 7143-a15/13 | 702 | 1 | 96 | 150 | 98.3 | 51 |
| 10-07-8383 | 2183 | 0.968 | 85 | 232 | 99.3 | 108 |
| 19-I-48528 A10 | 210 | 1 | 78 | 14 | 1.7 | 0 |
| 19-I-48528 A3 | 1534 | 0.324 | 123 | 629 | 99.4 | 122 |
| 430-2/10 | 2201 | 0.962 | 85 | 385 | 99.5 | 109 |
| PU14209-A4/17 | 2957 | 0 | 92 | 197 | 98.9 | 95 |
| PU1859-A1/15 | 5374 | 0 | 101 | 433 | 99.4 | 119 |
| PU4940-A2/12 | 1941 | 0.986 | 94 | 273 | 99.3 | 110 |
| CTRL-DNA | 30291 | 0.272 | 133 | 932 | 99.4 | 124 |
Table 2: DNA samples QC metrics. The table shows whether the processed samples met the required quality metrics for proceeding with downstream analysis. All samples meet minimum quality requirements, except for DNA sample 19-I-48528 A10, underlined in yellow, which has not been considered for SNV, TMB and MSI evaluation.
| Samples | QC metrics for fusion and splice | ||
|---|---|---|---|
| Median CV gene 500X (%) | Total on target reads (%) | Median insert size (Count) | |
| Lower Limits (LSL) Guidelines | 0 | ≥ 9000000 | ≥ 80 |
| Upper Limits (USL) Guidelines | ≤ 93.00 | NA | NA |
| PG2057-A4/17 | 54.4 | 13904129 | 105 |
| PG28902-A3/18 | 53.05 | 16921802 | 118 |
| PG5315-A4/14 | 65.52 | 15022172 | 98 |
| PU4215-A1/12 | 62.71 | 16864002 | 103 |
| PU5741-A9/17 | 61.06 | 12292999 | 99 |
| PU7055-A2/14 | 73.58 | 18672060 | 95 |
| PU7307-A3/13 | 56.32 | 14297184 | 109 |
| 11081-7/08 | 53.91 | 5800832 | 112 |
| 15613-3/08 | 54.49 | 17333112 | 97 |
| 20-I-59397 A5 | 43.53 | 13775359 | 130 |
| 7143-a15/13 | 57.96 | 17837144 | 96 |
| 10-07-8383 | 73.81 | 18016714 | 82 |
| ‘ 19-I-48528 A10 | 48.84 | 20220306 | 122 |
| 19-I-48528 A3 | 48.85 | 18972072 | 131 |
| 430-2/10 | 62.46 | 23311146 | 96 |
| PU14209-A4/17 | 54.84 | 19829529 | 102 |
| PU1859-A1/15 | 67 | 19815603 | 88 |
| PU4940-A2/12 | 64.5 | 23182096 | 92 |
| CTRL-RNA | 54.22 | 16600101 | 164 |
Table 3: RNA samples QC metrics. The table shows whether the processed samples met the required quality metrics for proceeding with downstream analysis. All samples meet minimum quality requirements in RNA specimens.
We have decided to test robustness of TSO-500 pipeline including one commercial DNA control (Multiplex gDNA Reference Standard, Horizon, REF: HD753) and one commercial RNA control (Universal Human Reference RNA, Agilent, REF: #740000) for each sequencing run performed. To increase the confidence of the obtained data we compared the mutational profile of the DNA and RNA controls throughout three different sequencing runs. DNA control sample was evaluated for the reproducibility of mutations VAFs detected, calculating mean values and standard deviations. Data for variants with low VAF are reported in Table 4. Selected variants were highly reproducible, with a mean standard deviation of 0.009. Tumor Mutation Burden and Microsatellite Instability values were also compared among each sequencing run and the reproducibility of data was confirmed, with data showing small standard deviation from mean values, as reported in Table 5.
| Sample variant | Allele frequency | Statistic | ||||
|---|---|---|---|---|---|---|
| Gene | Variant | RUN I | RUN II | RUN III | MEAN | ST. DEV. |
| GNA11 | Q209L | 0,0692 | 0,0448 | 0,0347 | 0,050 | 0,018 |
| AKT1 | E17K | 0,0480 | 0,0430 | 0,0376 | 0,043 | 0,005 |
| PIK3CA | E545K | 0,0460 | 0,0527 | 0,0571 | 0,052 | 0,006 |
| EGFR | p.(Ala767_Val769dup) | 0,0382 | 0,0292 | 0,0345 | 0,034 | 0,005 |
| EGFR | ΔE746-A750 | 0,0431 | 0,0498 | 0,0299 | 0,041 | 0,010 |
Table 4: DNA control evaluation: VAF reproducibility. HD753 DNA control analyzed for detected variants VAF reproducibility. Mean values and standard deviations are reported in the table.
| RUN I | RUN II | RUN III | MEAN | ST.DEV | |
|---|---|---|---|---|---|
| TMB | 307,2 | 312,1 | 311,8 | 310,37 | 2.75 |
| MSI (%) | 67,20 | 67,74 | 70,16 | 68,37 | 1.58 |
Table 5: DNA control evaluation: TMB and MSI reproducibility. HD753 DNA control analyzed for Tumor Mutation Burden and Microsatellite Instability reproducibility. Mean values and standard deviations are reported in the table.
RNA control sample was tested for the reproducibility of the rearrangements declared to be present form the manufacturers throughout all the three performed sequencing runs. Every rearrangement was detected in each run, at the same genomic location and with the same fusion partners. The passing criteria for eligibility of the rearrangement were met from all the fusions detected throughout each run and the score assigned to each fusion was reproducible and indicate the quality of the detected rearrangement. The higher the score, the more realistic the detected fusion is. Reproducibility of commercial RNA control is reported in Table 6.
| Fusion partner 1 | Fusion partner 2 | Genomic location partner 1 | Genomic location partner 2 | RUN I | RUN II | RUN III | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pres. | Score | Pres. | Score | Pres. | Score | PASS | ||||
| ESR1 | CCDC170 | chr6: | chr6: | ٧ | 0.632 | ٧ | 0.6 | ٧ | 0.612 | PASS |
| 152023138 | 151894307 | |||||||||
| WHSC1L1 | FGFR1 | chr8: | chr8: | ٧ | 0.716 | ٧ | 0.7 | ٧ | 0.703 | PASS |
| 38239316 | 38315051 | |||||||||
| RPS6KB1 | VMP1 | chr17: | chr17: | ٧ | 0.724 | ٧ | 0.8 | ٧ | 0.849 | PASS |
| 57987972 | 57915656 | |||||||||
| RPS6KB1 | VMP1 | chr17: | chr17: | ٧ | 0.693 | ٧ | 0.7 | ٧ | 0.799 | PASS |
| 57992062 | 57917127 | |||||||||
Table 6: RNA control evaluation: Rearrangements reproducibility. Universal Human Reference RNA control analyzed rearrangements reproducibility. Fusions reported to be present in the sample from manufacturers were present in each sequencing run at the same genomic location for both fusion partners and with reproducible scores.
Data collection and PierianDx genomic workspace interpretation
Primary and secondary analysis were performed in BaseSpace, a cloud-based, Illumina propriety database where users can store, analyze and share data. Sequencing data generated by Illumina sequencers are stored in real-time into this software and secondary analysis, after FASTQ generation, can be done. Primary analysis is the process of elaborating raw sequencing data, namely Binary Base Call (BCL) files, into nucleotide base and short-read data (FASTQ) [28]. During the secondary analysis, the FASTQ generated files are aligned to hg19 reference sequence with subsequent variant detection and annotation [28]. For the tertiary analysis, involving data interpretation and reporting, we took advance of the collaboration between Illumina and Pierian Dx Clinical Genomics Workspace (CGW). This company provides clinically integrated software aimed to the interpretation of genomic data and delivery of genomic insights for a precise and accurate patient care. Pierian DX knowledgebase it’s based on powerful rule engine delivering rationalized medical interpretations to sequencing data, together with expertly curated genomic data and curated sources, such as NCCN/ASCO guidelines, FDA therapies, clinical trials and published literature. Furthermore, all shared genomic interpretations in the database are curated by clinical customers and interpretation services team, with an acute focus on their impact for clinical care [29]. This collaboration between PierianDx and Illumina gives access to the laboratories to a powerful tool providing clients and clinicians with a complete and specific report, allowing clinicians to discuss the clinic and the therapeutic applications of the genetic data for a better and targeted patient management.
After secondary analysis, we provided Pierian Dx with the following set of files for the data interpretation and report generation: dsdm/json file, containing all the executed analytical steps; the sample sheet in csv format; any information related to the exact tumor type that we were evaluating; DNA sample Merged Small Variants vcf file, containing a report with all the annotated single nucleotides variations; DNA sample copy number variants vcf file; DNA sample tmb file in json format and the DNA sample msi file in json format, to evaluate tumor mutation burden and microsatellite instability biomarkers; RNA sample all fusions file in csv format and RNA sample splice variants file in vcf format.
Reports were generated using the following approach. Variants were included in the report using an arbitrary cut-off of 5% [30]. No variants were included with a variant allele frequency less than 5%. Variant population frequency was considered for the classification. Somatic variants are reported and listed according to TIER classification [31]. Tier III VUS variants were added to the report if the gnomAD Total Population Frequency was less than 0.5% irrespective of variant fraction. For each type of tumor, the presence of any ongoing clinical trials was taken into consideration, thus making it possible to classify some of the variants identified as Tier IIC variants. For the evaluation of TMB the following thresholds were set: a threshold of 5 mutations per MB as low TMB, 5-10 mutations per MB as medium TMB and >10 mutations per MB as high TMB [32,33]. The threshold for MSI evaluation was set at >20% for unstable Microsatellite Instability (MSI) [34]. Variant classification was changed from the one given automatically by the CGW platform in order to resolve ambiguities, reflect variant population frequency or the presence of potentially suitable clinical trials.
The analysis of this study were carried on considering only the mutations included in the following classes: Tier I A, referring to variants of strong clinical significance with level A evidence, meaning FDA approved therapy or practice guideline in patient's tumor type; Tier I B, referring to variants of strong clinical significance with level B evidence, meaning consensus in the field based on well powered studies in patient's tumor type; Tier II C, referring to variants of potential clinical significance with level C evidence, meaning FDA approved therapy or practice guideline in other tumor types, evidence from multiple small published studies, or based on availability of investigational therapies; Tier II D, referring to variants of potential clinical significance with level D evidence, meaning found in case reports or preclinical studies; Tier III, referring to Variant of uncertain clinical significance.
Variants belonging to these tiers were selected by Pierian Dx to be included in the clinical report, since we want to evaluate data from a clinical point of view and data which are therapeutically relevant for affected patients.
Mutational landscape of PDTC and ATC: A comparison with literature
In this preliminary study we analyzed 10 poorly differentiated thyroid carcinoma and 8 anaplastic thyroid carcinoma samples using a comprehensive genomic profile panel produced by Illumina. The TSO-500 platform analyzes, with a hybrid capture-based technology, 523 genes related to oncogenesis and oncological diseases. We compared the somatic mutational landscape of our study cohort with the one reported in literature, to evaluate if the mutational footprint of PDTC and ATC was reflected also in our study settings.
It is known from previous WGS and WES studies that PDTC and ATC present different driver mutations, defined by Julia R. Pon and Marco A. Marra [35] as genomic variants providing a selective growth advantage and therefore promoting cancer development. Driver mutations mainly involves RAS-RAF-MAPK and PIK3-AKT pathways, or interest cell cycle regulation, chromatin remodeling, DNA damage response and protein metabolism functions [12,14]. The most well characterized driver mutations in PDTC and ATC are BRAFV600E and RAS (KRAS, NRAS and HRAS) mutations [12,14]. According to a 2016 study aimed at describing the genomic and transcriptomic profile of PDTC and ATC [16], BRAFV600E mutations were present in 33% of PDTC analyzed cases and 45% of ATC analyzed cases, whereas mutations in NRAS, HRAS, or KRAS occurred in 28% and 24% of PDTCs and ATCs, respectively. According to literature and to COSMIC database [36], mutations in PI3K-AKT intracellular signaling pathway are more relevant in ATC compare with PDTC cases, with PIK3CA and PTEN mutated in 11, 24% and 9.24% of cases respectively. Even if ATCs are reported to be more likely mutated in PI3K-Akt pathway, PDTC samples also show 2.38%-19.51% of mutations in PIK3CA and 0-8.70% in AKT1.Together with mutations at intracellular signaling pathway levels, also cell cycle regulation actors has been shown to be highly mutated in PDTC and ATC cases, with p53 and TERT promoter mutations being more frequent in ATC (45, 67% and 75%) when compared with PDTC cases (8% and 40%) [12,37].
Chromatin remodeling pathway mutations are also known for their tumorigenic potential. Chromatin remodeler proteins determine the accessibility of different factors to nucleosome DNA, allowing the activation or the repression of several pathways and a whole repertoire of biological functions. For this reason, genetic variants in genes involved in chromatin remodeling pathway leading to aberrant expression or epigenetic modulation, drives cancer cells to reprogram their genome for the maintenance of oncogenic phenotypes [38]. NGS data from literature indicates a major contribute of chromatin remodeling mutations in tumorigenesis of ATCs, while not relevant data are found for what concern PDTC cases [12]. Also protein metabolism pathway is known to be involved in tumorigenesis [39]. According to literature, both PDTC and ATC harbor EIF1AX mutations in 10% of cases [12].
In Figure 3 we report and compare the percentage of patients harboring mutated genes between PDTC and ATC samples of our study cohort, considering as eligible variants all single nucleotide variants, splice variants and small indels classified by Pierian Dx as Tier I A, IB, IIC, IID and Tier III. Genes under analysis were subdivided according to the biological pathways of belonging. Our data approximately resemble the mutational landscape found in literature, but our statistic suffers from the low number of samples of this study cohort.
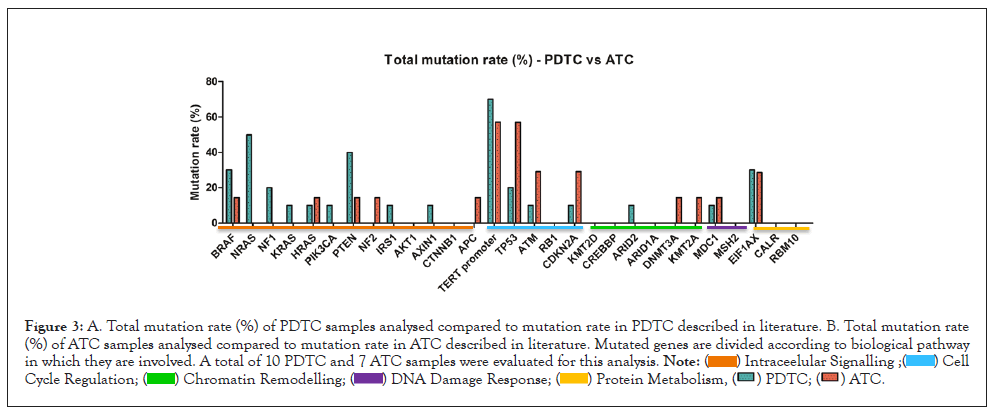
Figure 3: Figure 3: Total mutation rate (%) indicates the percentage of patients in which the analyzed gene harbored Tier I, II or III variants. Here we compare the total mutation rate (%) of PDTC samples analyzed to the total mutation rate (%) of ATC samples analyzed. Mutated genes are divided according to biological pathway in which they are involved. A total of 10 PDTC and 7 ATC samples were evaluated for this analysis.
Nevertheless, we can speculate that mutations at cell cycle regulation level seems to be affecting more ATC samples when compared with PDTCs, as reported in literature, with a higher mutation rate detected in ATM and CDKN2A genes in ATC samples when compared with data from previous studies (Figure 4) [12]. Together with this, a higher contribution of chromatin remodeling dysfunctional pathway in ATC tumorigenesis compared with PDTC can also be seen. BRAF and RAS genes seem to be highly mutated in both PDTC and ATC samples. Literature states that mutations in PI3K-AKT intracellular signaling pathway are more relevant in ATC compare with PDTC cases [12], but our study reports higher mutation rate in PTEN, NRAS and NF1 genes for PDTC than for ATC sample, and higher mutation rate than data reported in literature (Figure 4).
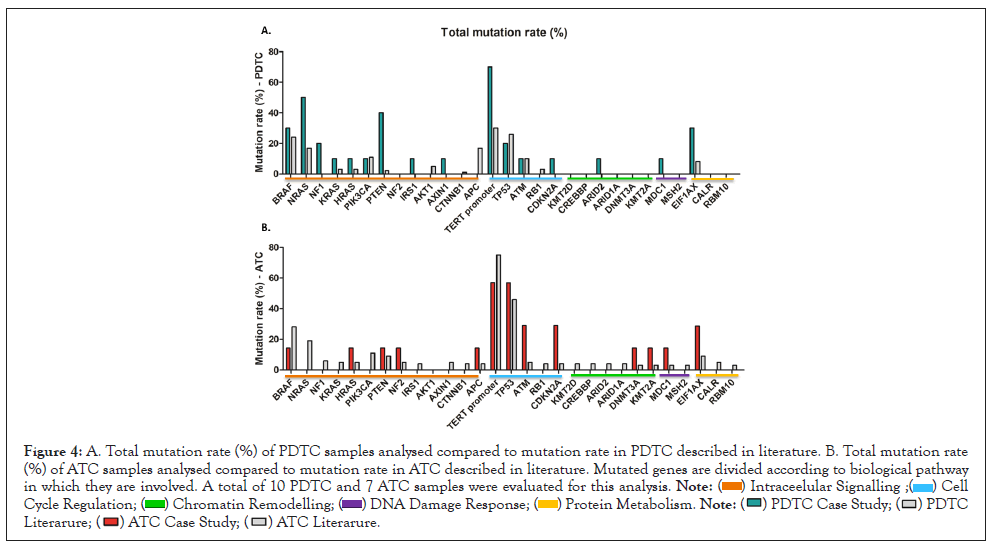
Figure 4: A. Total mutation rate (%) of PDTC samples analysed compared to mutation rate in PDTC described in literature. B. Total mutation rate (%) of ATC samples analysed compared to mutation rate in ATC described in literature. Mutated genes are divided according to biological pathway in which they are involved. A total of 10 PDTC and 7 ATC samples were evaluated for this analysis.
Due to the low sample number of this preliminary study, differences seen in the mutation rate of these different thyroid carcinomas should be carefully interpreted and rationalized and more deeply investigated, increasing the ATC and PDTC cohort. Protein metabolism related genes, and in particular EIF1AX gene, seems to be highly mutated both in PDTC and ATC samples, as reported in literature [12]. As shown in Figure 4, higher mutation rate is shown in our study when compared with values reported in previous studies. We detected 28,57% mutation rate for EIFAX gene in ATC samples and 30% mutation rate in PDTC.
As stated in paragraph 3.1, ATC sample 19-I-48528 A10 was not considered for the SNV evaluation because of bad quality metrics results for DNA sample.
Mutational landscape of PDTC and ATC and tumor profiling
We also investigated the mutational landscape of our sample cohort to evaluate the ability of TSO-500 platform to differentiate and to profile different tumor types according to their mutational signature. Profiling data are shown in Figure 5. Mutation frequencies are similar throughout all the different cellular pathways. Nevertheless, we can discriminate an increased mutational rate at intracellular signaling pathway level for PDTC when compared with ATC. The possibility of this analytic pipeline to differentiate tumor types according to their different mutation profile is of main importance because it can potentially help clinicians in choosing the more appropriate and personalized therapy for the affected patient. Even if the cancer type is unknown or morphologically ambiguous as in case of PDTC and ATC samples, a molecular characterization of mutational landscape might be helpful for a better patient profiling and stratification for therapies and treatment.

Figure 5: Each column represents an individual tumor. Genes are sorted according to the biological pathway to which they belong, as intracellular signaling, cell cycle regulation, chromatin remodelling, DNA damage repair, protein metabolism, cell migration, adhesion and motility, cell proliferation. The right bar chart represents the frequencies of gene alterations across 10 PDTC and 7 ATC samples.
As stated in paragraph 3.1, ATC sample 19-I-48528 A10 was not considered for the mutational landscape evaluation because of bad quality metrics results for DNA sample.
The mutation burden rate along the spectrum of thyroid carcinomas progression
Tumor Mutation Burden, or TMB, is defined as the number of somatic mutations per Mb of interrogated genomic sequence [24]. Tumor-specific neoantigens arise from somatic mutations and can increase the likelihood of immune recognition and tumor cell killing. The immune system is in fact stimulated against these tumor neoantigens. TMB has therefore been widely explored as an alternative or complementary biomarker for response to Immune Checkpoint inhibitor therapies [40].
WES is generally considered as the definitive standard for TMB calculation, but the use of DNA large panels covering 1.1 Mb or more of genomic content [40] is an emerging opportunity to have a good TMB estimation [41]. Good correlation between TSO500 and WES is established [42].
For what concern TMB and thyroid carcinomas, it has been shown to correlate with the degree of differentiation tumor presents, with an increase of TMB along the spectrum of Thyroid Carcinoma Progression. The median mutation burden detected in PTC, PDTC, and ATC is 1, 2, and 6 per tumor, respectively [14]. Our preliminary data confirm this trend significantly. Mann Whitney test was used to assess significance between ATC and PDTC TMB values. TMB values, as shown in Figure 6, are significantly higher for ATC samples compared with PDTC samples with a p value of 0.0212.
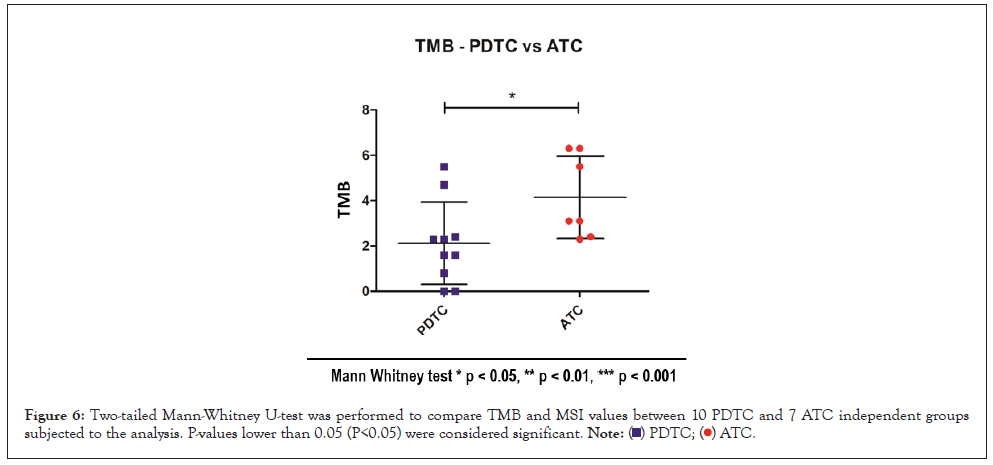
Figure 6: Two-tailed Mann-Whitney U-test was performed to compare TMB and MSI values between 10 PDTC and 7 ATC independent groups subjected to the analysis. P-values lower than 0.05 (P<0.05) were considered significant.
As stated in paragraph 3.1, ATC sample 19-I-48528 A10 was not considered for the TMB evaluation because of bad quality metrics results for DNA sample.
Correlation between TMB and MSI
Microsatellite instability, or MSI, is a form of genomic instability resulting in the accumulation of insertions or deletions (indels) in microsatellites during replication due to a dysfunctional mismatch repair (MMR) system. MMR proteins are responsible for correcting errors made by DNA polymerase during replication. They do so by recognizing a temporary insertion-deletion loop that is created when DNA polymerase slips. Cells with a dysfunctional MMR protein accumulate errors when the loop resulting in frameshift mutations (indels), leading to the appearance of novel alleles at microsatellite loci, which can be easily identified via fragment analysis. In addition to screening for cancers, MSI has recently been found to be predictive of response to immunotherapies. Tumors with defective MMR proteins often have somatic cells with mutations that produce novel proteins that can be immunogenic [25].
MSI is traditionally analyzed with PCR (MSI-PCR). However, NGS allows for the analysis of a greater number of microsatellite loci than MSI-PCR, with high concordances with MSI-PCR technique, presenting opportunities to identify new MSI profiles in previously uncharacterized cancer types [40]. The microsatellite instability status is calculated reporting stable or instable sites for evidence of instability relative to a set of baseline normal samples.
Some papers have investigated the correlation between TMB and MSI in solid tumors [43,44]. Coupling MSI and TMB analysis, may represent a decisive tool for selecting patients for immunotherapy, for common or rare cancers. We were curious to investigate this potential combined biomarker [43,44]. Upon statistical analysis, no correlation between TMB and MSI could be detected in ATC samples while in PDTC a significative correlation could be identified (Figure 7), with high TMB values corresponding to high MSI status. As already stated, this data might suffer from the poor sample numbers and have to be carefully interpreted.
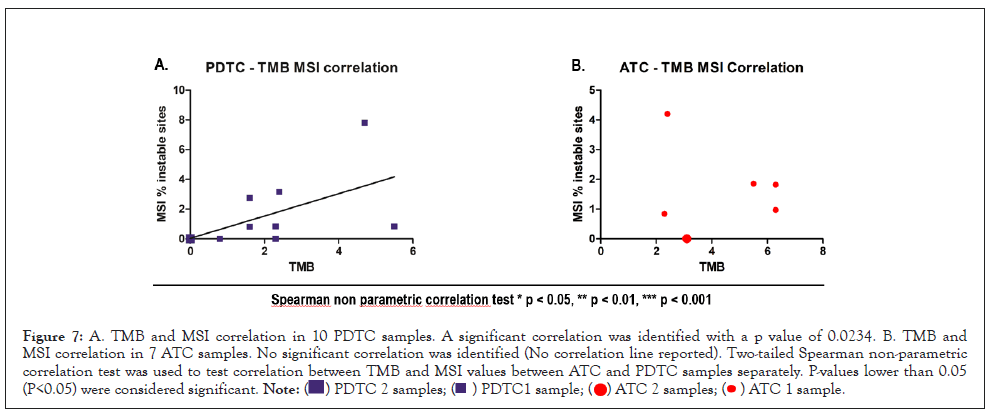
Figure 7: A. TMB and MSI correlation in 10 PDTC samples. A significant correlation was identified with a p value of 0.0234. B. TMB and MSI correlation in 7 ATC samples. No significant correlation was identified (No correlation line reported). Two-tailed Spearman non-parametric correlation test was used to test correlation between TMB and MSI values between ATC and PDTC samples separately. P-values lower than 0.05 (P<0.05) were considered significant.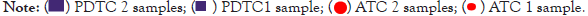
As stated in paragraph 3.1, ATC sample 19-I-48528 A10 was not considered for the TMB and MSI correlation evaluation because of bad quality metrics results for DNA sample.
Fusion analysis and validation
RNA fusion analysis was carried out on two ATC samples, in which 3 different rearrangements were found from the TSO-500 pipeline. All fusions were found between NTRK3 and ETV6 genes. The fusion involving these two genes is common in TCs, representing the second most common rearrangement seen in the post-radiation setting [45]. The ETV6 gene expresses a transcription factor protein, ETV6, required for hematopoiesis and usually involved in malignant transformation. NTRK3 encodes a member of the Neurotrophic Tyrosine Receptor Kinase (NTRK) family, a cell surface protein that when bound to its growth factor ligand stimulates signaling proteins that promote the growth, survival, and proliferation of the cells. The tyrosine kinase of the ETV6-NTRK3 fusion protein is dysfunctional and therefore it is continuously active in phosphorylating tyrosine residues on, continuously stimulating proteins that promote the growth, survival, and proliferation of their parent cells [46].
In order to validate the presence of these 3 identified fusions, other detection techniques should be performed on the analyzed samples, as FISH or Sanger sequencing with primers designed specifically for the detected rearrangement. We decided to design rearrangement specific primers and validate the fusions via Sanger Sequencing. Extrapolating the sequence of these 3 rearrangements from the data generated from the Basespace secondary analysis, we could design fusion specific primers and verify their presence. Due to poor primer specificity we were able, up to now, to isolate and sequence only one of the three fusions. The sequence of the fusion and breakpoint regions were confirmed comparing sanger electropherogram with the sequence generated during the secondary analysis. We also decided to Blast [47] ETV6 and NTRK3 partner to confirm the fusion partner calls generated by the secondary analysis. After collecting all the data, the identified fusion was validated. The sequence and the primer sequence of the identified region are reported in Figure 8.
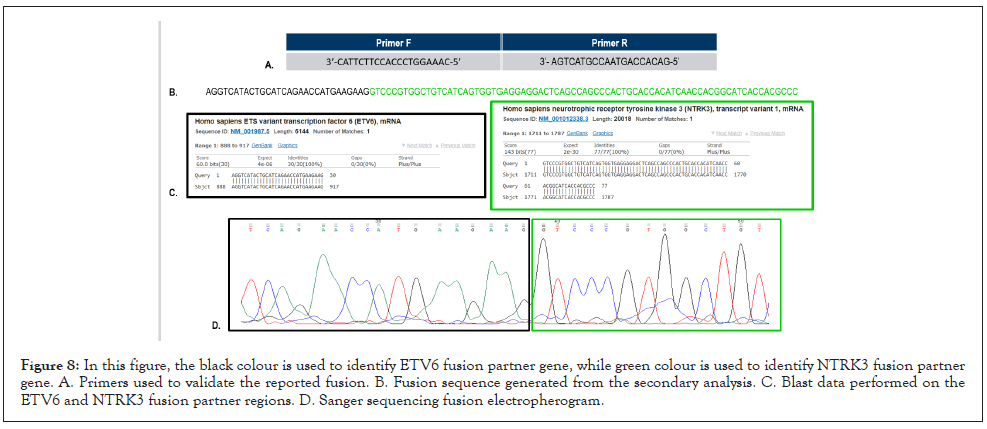
Figure 8: In this figure, the black colour is used to identify ETV6 fusion partner gene, while green colour is used to identify NTRK3 fusion partner gene. A. Primers used to validate the reported fusion. B. Fusion sequence generated from the secondary analysis. C. Blast data performed on the ETV6 and NTRK3 fusion partner regions. D. Sanger sequencing fusion electropherogram.
Poorly differentiated and anaplastic thyroid carcinomas are rare but aggressive thyroid tumors. Their characterization has been challenging since the high degree of dedifferentiation that these cancers present. Need for a better characterization and the urge to have a quick diagnosis to provide treatment to these short life expectancy patients, led in the last decades to improve the molecular characterization of these tumor types. This can help researchers and clinicians to find molecular biomarkers making the diagnosis more straightforward, to identify mutated biological pathways making the patient eligible for target therapies, and to identify biomarkers suggesting a type of response for the treatment with immune checkpoint inhibitors. For all these reasons we decided, in this study, to analyze 18 FFPE samples, 10 PDTCs and 8 ATCs, using a Comprehensive Genomic Profiling Approach (CGP), taking advance of the most recent technologies in NGS field.
Despite the number of advantages of NGS, this technique harbors some limiting factors. In the oncological field, the most utilized source of DNA or RNA for library preparation is FFPE samples and it is known that this fixation technique causes cross-linking and fragmentation of nucleic acids, resulting in low-quality DNA and RNA, leading to bad results [48]. Large volumes of data are acquired in NGS that need to be carefully evaluated to distinguish significant variants, from those generated from sequencing errors and background noise variations. A negative result therefore needs to be interpreted in the light of sample adequacy and read depth [48]. Moreover, traditional NGS techniques only evaluate single nucleotide variants and small indels, making it necessary to implement with other tests the analysis of difficult and rare cancer cases. It is mandatory, when analyzing data from different or rare tumor types, to take into consideration that one mutation might have different impact in different tumors, possibly being predictive in some tumors, but having no clinical implications in others. Therefore, also tumor heterogeneity remains a major issue with NGS based approaches, particularly when low frequency variants are found. These may represent subclones that could confer resistance to certain drugs [48].
The technical improvements in the NGS field and the necessity for having a clinical application of the retrieved data led to the generation of comprehensive genomic profiling assay. This approach makes it possible to evaluate in parallel known and unknown variants of the four main classes of genomic alterations, namely single nucleotide variants, indels, copy number variations and gene fusions, analyzing in one sequencing run both DNA and RNA extracted from FFPE samples. The panel evaluates a comprehensive set of cancer-relevant genes and reports complex biomarkers, such as TMB and MSI, indicative of responsiveness to immune-checkpoint inhibitor therapy. The results of the analyses are summarized in a clinical report, in compliance with ESMO guidelines, to provide prognostic, diagnostic and predictive insights that might implement and drive treatment decisions for patients across all cancer types.
TSO-500 is a Comprehensive Genomic Profile panel that analyzes 523 cancer-relevant genes from both DNA and RNA coming from FFPE tissue in one integrated workflow. The assays simultaneously assess multiple variant types from DNA and RNA samples, eliminating the need to spend precious tissue sample and time on iterative testing. The resulting panels provide comprehensive coverage of including 523 genes looking for Single Nucleotide Variants (SNVs), insertions/deletions (indels), Copy Number Variations (CNVs) and 55 genes for known and novel fusion and splice variants. In addition, the TSO-500 panels include the Microsatellite Instability (MSI) and Tumor Mutational Burden (TMB) analysis.
In this preliminary study we could assess assay robustness and reproducibility, performing library preparation and sequencing on one DNA and one RNA commercial control. Sequencing results were reproducible and showed comparable VAFs for the identified variants throughout all the three runs, for both starting materials.
Taking advantage of this innovative and performing approach, we were able to correctly profile rare and not yet completely characterized tumor types such as PDTC and ATC samples. Results of our study seem in fact to recapitulate pretty well the PDTC and ATC profiling already reported in previous studies. Considering our small sample cohort, we still can see that mutation rates of our samples approximately resemble the mutational landscape found in literature, with high number of mutations affecting mostly genes of the intracellular signaling and cell cycle regulation pathway. We depicted some outliers from this analysis. In our PDTC sample set in fact, we can depict a higher number of patients carrying mutations in NRAS, NF1 and PTEN gene compared with the rate reported in literature. In ATC settings instead, data show a higher number of patients carrying mutations in ATM and CDKN2A cell cycle regulation gene when compared with literature. Lastly, we identified higher mutation rates in protein metabolism gene EIF1AX, both in PDTC and ATC samples, when compared with data retrieved in literature.
NRAS mutations are quite common both in PDTC and ATC samples, co-occurring with BRAF and other RAS family mutations. On the other side NF1 and PTEN mutations were found mainly in ATC samples in previous studies, but not in PDTCs [16]. For this reason, a correlation between increased NRAS, PTEN and NF1 mutated gene might be of interest for further investigations. ATM is a cell-cycle regulator and a DNA damage response gene which is shown to be highly mutated in our samples. Previous studies report a correlation between mutated ATM gene and high mutational burden values [16]. It might be intriguing to evaluate this correlation with a wider case of samples. EIF1AX gene encodes for a protein required for increasing the rate of protein biosynthesis, enhancing ribosome dissociation into subunits and stabilizes the binding of the initiator Met-tRNA(I) to 40 S ribosomal subunits [49]. EIF1AX has been previously reported to be mutated in approximately 10% of cases of PDTC and ATC cases, with a strong concordance with RAS family mutations [16]. The biological consequences of this association are currently unknown, but we can appreciate it in our sample. Together with this, the increased mutation rate that we observe in our dataset compared with literature data makes EIF1AX gene an interesting target for further investigations.
Our preliminary study allowed us to depict a tumor specific profile, with tumor specific signatures that vary from PDTC to ATC samples. PDTC environment is showed to be more highly mutated at intracellular signaling pathways, while lower mutation rate at this level can be identified in ATC samples. We were also seeing a similar differential profile involving DNA damage response pathway. On the other hand, there is a trend for higher mutation profile in genes involving chromatin remodeling pathway for ATC when compared with PDTCs. By increasing sample cohort, we might be able to better stratify PDTC and ATC affected patients and might be able to also stratify patients with difficult and non-conclusive diagnosis with profiles resembling the one already analyzed, opening the way to new and out of label treatment options that might improve patient performances and clinical outcome. This approach in fact potentially helps clinicians in evaluating personalized therapy for the affected patient because a molecular characterization of the mutational landscape helps to profile a complex diagnosis patient, allowing stratification for therapies and treatment.
Together with this, TSO-500 provides also accurate data regarding tumor mutation burden and microsatellite instability, reflecting data found in literature, proving assay robustness and efficacy in identifying relevant biomarker which might help clinicians to evaluate therapeutic strategies for patient. Finally, in our preliminary study we were able to validate the presence of fusions in one ATC sample, proving the reliability of the CGP approach in identifying both DNA and RNA variations for a better patient genomic landscape characterization.
Illumina CGP approach offers in fact a complete evaluation of the genomic landscape of each tumor for both prognostic and predictive purposes, helping oncologists in making decisions about cancer treatment and may support the administration of immunotherapy drugs in patients harboring high TMB. The main, remarkable advantage of CGP in the diagnostic field is that this approach consolidates biomarker detection into a single multiplex assay, eliminating the need for sequential testing, making it possible to assess the most prevalent as well as rare biomarkers. This approach increases the chances of finding a positive biomarker, providing faster results, limiting the input of precious biopsy samples. As a results, this comprehensive approach translates into shorter report times meanwhile increasing the screening yield. Without such a comprehensive approach evaluating DNA and RNA in parallel we would not have been able to characterize fusions, resulting into a lack of information and a worst impact on patient profiling. As remarkable consequence, this comprehensive genome panel approach allows for shorter response times, which translates into the possibility to evaluate quickly available and responsive therapeutic options for the patient. In addition to that, this also opens the doors to a better screening options, better surveillance, and more accurate screening procedures to patient relatives, providing in the end a better outcome for the patient and patient family. In this context, comprehensive profiling approach might also be useful to characterize more in depth those variants in genes whose contribution to cancer risk modulation is not completely depicted yet. In fact, within our experience with TSO-500 validation, we identified potentially interesting variants in different colorectal cancer samples that are currently being investigated with population and functional studies. These variants have been identified with higher frequencies when compared with general population data and their functional role has not been elucidated yet. Their recurrency led us to think of them as potentially involved in cancer predisposition or development. If so, this could result in the identification of novel biomarkers relevant for risk identification and patient stratification, helping clinicians in the planification of cancer prevention strategies and in personalized patient care. In this context, comprehensive profiling approach might also be useful to characterize more in depth those variants in genes whose contribution to cancer risk modulation is not completely depicted yet. In fact, within our experience with TSO-500 validation, we identified potentially interesting variants in different colorectal cancer samples that are currently being investigated with population and functional studies. These variants have been identified with higher frequencies when compared with general population data and their functional role has not been elucidated yet. Their recurrency led us to think of them as potentially involved in cancer predisposition or development. If so, this could result in the identification of novel biomarkers relevant for risk identification and patient stratification, helping clinicians in the planification of cancer prevention strategies and in personalized patient care.
In conclusion, we believe that Illumina TSO-500 approach coupled with Pierian Dx reporting services, offers an accurate and effective approach for PDTC and ATC cases evaluation, offering clinicians reliable solution for patient evaluation. The wide panel design allows the simultaneous evaluation of a broad range of genetic variations and clinical biomarkers allowing a fast and complete patient genomic profiling. This will help clinicians in evaluating best treatment options for not fully characterized cancer cases, according to the genomic landscape of analyzed cases.
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar] [PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
Citation: Dellai A, Scandurra A, Moretti S, Mandarano M, Sidoni A, Puxeddu E, et al. (2022) Genomic Profiling in Anaplastic and Poorly Differentiated Thyroid Carcinomas: A Preliminary Study. J Clin Chem Lab Med. 5:226.
Received: 16-Jun-2022, Manuscript No. JCCLM-22-17963; Editor assigned: 20-Jun-2022, Pre QC No. JCCLM-22-17963 (PQ); Reviewed: 04-Jul-2022, QC No. JCCLM-22-17963; Revised: 11-Jul-2022, Manuscript No. JCCLM-22-17963 (R); Published: 18-Jul-2022 , DOI: 10.35248/JCCLM.22.05.226
Copyright: © 2022 Dellai A, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.