Mycobacterial Diseases
Open Access
ISSN: 2161-1068
ISSN: 2161-1068
Research Article - (2020)Volume 11, Issue 6
Background: Rapid diagnosis of TB globally remains challenging and emergence of resistant strains represents a significant threat. Improved techniques on diagnosis of tuberculosis is a major concerning for this burden control and is crucial for new tuberculosis control strategy. This study aimed to evaluate diagnostic accuracy of GeneXpert MTB/RIF assay for confirming rifampicin resistance among those suspects comparing to multiplex PCR, RFLP-PCR and real-time PCR amplifications.
Methods: Sputum samples were collected and analysed during 2021 to 2023. Sputum was transferred to the bacteriology laboratory for MTB/RIF detection using conventional bacteriological and molecular based-assays. Sensitivity and specificity were accounted for the stained smear and other molecular methods according to GeneXpert MTB/RIF as the gold standard.
Results: All 93 samples were analyzed for rifampicin resistance by GeneXpert MTB/RIF compared to others, of 73 (78.49%) positive samples, 59 (63.44%) and 17 (18.27%) were positively reported as susceptible and resistance respectively, while 17 (18.27%) negatively reported. The positivity rates, PPV and NPV for microscopy (77.41%, 87%, 76%), real time PCR (78.49%, 84%, 72%), RFLP-PCR (78.49%, 80%, 85%) and multiplex PCR (67.74%, 78%, 56%) respectively. Among those molecular diagnostic techniques, only RFLP-PCR followed by GeneXpert results with the sensitivity and specificity of 96% and 100%, respectively, was reported as the most powerful method compared to others for diagnosis of Mtb and the genome sequence of rifampicin resistance.
Conclusions: Both modern and conventional molecular techniques can be aligned and used together for rapid and conclusive diagnosis of Mtb and rifampicin resistance region, rpoB.
GeneXpert MTB/RIF; Mycobacterium tuberculosis; Drug resistance
Tuberculosis is still a serious public health problem and healthcare systems face with a significant diagnostic and therapeutic challenge globally. Using diverse molecular diagnostic techniques play key roles in the World Health Organization’s (WHO) new tuberculosis control strategy. WHO reported 10.4 million new cases of tuberculosis and 1.7 million deaths (including 0.4 million co-infected with HIV) in 2016. In case of anti-bacillary resistance, WHO reported a rifampicin resistance rate of 3.5% in new diagnosed TB cases whereas 18% in treated patients. In Iran, in 2019, there was an estimated of TB incidence of 13 per 100,000 populations. Also, MDR/ Rifampicin (RIF)-resistant TB rates were 1.3% among new cases with an estimation of 8.3% in retreatment cases. Patients diagnosed with RIF-resistant, as known for proxy MDR-TB, mostly need longer and ineffective treatments compared to firstline procedures. The most laboratories are unequipped for Drug Susceptibility Testing (DST) in Iran, causing the diagnosis of RIF-resistance fairly poor in the country. Hence, RIF-resistant-TB, very often remains unsolved, making more spread of drugresistant TB cases and incomplete TB treatment outcomes. Bearing in mind that RIF-resistant TB is among the major threat for national TB Control Programs (NTP), detecting RIFresistant TB resistance among Mycobacterium tuberculosis isolates will make a better advance treatment achievement [1].
Currently, conclusive detection of tuberculosis and anti-bacillary resistance relies on the conventional laboratory methods that require the bacterial growth of different dilutions of mycobacteria in solid or liquid media in the presence of known concentrations of anti-bacillary. However, these methods have drawbacks such as requiring expert technicians, the need for high biosafety levels and the time delay in reporting. Conversely, the adoption of molecular diagnostic tests for anti-bacillary resistance makes it possible to reduce the time-to-results and additively increases the diagnostic sensitivity by directly searching for mutations in the genes that determine resistance to different anti-bacillary. Different molecular tests are GeneXpert MTB/RIF (cepheid, sunnyvale, CA, USA) and the reverse hybridization test on strips genotype MTBDR plus (HAIN life sciences, Nehren, Germany). GeneXpert MTB/RIF is now the only rapid molecular test announced by WHO for the rapid diagnosis of tuberculosis. GeneXpert MTB/RIF can detect Mtb complex genome in patient specimens and identify the genome responsible of the main mutations versus rifampicin resistance (rpoB gene mutation). MTBDRplus is a kind of multiplex DNA PCR test coupled with hybridization on strips for simple identification of mycobacteria and detection of genomic sequences of anti-tuberculosis drug resistance. The result can be found in a few hours and rapid identification of anti-TB like rifampicin and isoniazid resistance can also be detected by a single amplification test. Using different molecular techniques for conclusive identification of tuberculosis is considered by WHO for making comprehensive tuberculosis control strategies. Some of current reports on the performance of GeneXpert MTB/RIF and genotype MTBDRplus in detecting rifampicin resistance have shown different results that are mainly relied on the regions at which those trials were performed.
The objective of this study is to evaluate the performance of GeneXpert MTB/RIF (cepheid, sunnyvale, CA, USA) in the diagnosis of Mtb and the genome sequence of rifampicin resistance, rpoB gene mutations among the suspects comparing to multiplex PCR, Restriction Fragment Length Polymorphism-PCR (RFLP-PCR) and real-time PCR amplifications [2].
This cross-sectional study included of 93 sputum samples of suspected patients infected with TB from 2021 to 2023. The samples were collected and transferred to the central bacteriology laboratory of Massih Daneshvari hospital, referral tuberculosis research center in Tehran, Iran for diagnosis of Mtb by bacteriological methods including microscopy and other molecular based techniques such as GeneXpert MTB/RIF, RFLP-PCR, multiplex PCR and real-time PCR. The study protocol was approved by the local ethics committee of faculty of medicine, Massih Daneshvari hospital and all the patients and/or their guardians have signed informed consent.
Acid-Fast Bacillus (AFB) smear microscopy
The samples were first stained using Ziehl-Neelsen on nondecontaminated sputum samples (liquid samples were concentrated for 15 min at 3000 rpm and sediments were stained). Prior to smear preparation, purulent sputum was mixed with N acetyl-L-cysteine as a mucolytic agent to enhance the homogeneity of the sample. Smears were then examined to confirm the presence of acid-fast bacilli and arranged as per the international union against tuberculosis and lung disease scale; negative for TB, +1, +2 and +3 and more. A patient was considered positive if a minimum of one smear was graded.
GeneXpert MTB/RIF diagnostic system
The GenXpert MTB/RIF protocol was used based on the manufacturer structure of GenXpert TMMTB/RIF, Cepheid (2009 April). In brief, the proceeded samples were diluted with Sample Reagent (SR) at a ratio of 1:2. The sample reagent mixture was then shacked for at least 10 s and incubated at room temperature for a total of 10 min. Next, the digested mixture was sent to the GenXpert MTB/RIF cartridge and the automated steps of the procedure were started immediately after adding the sample to the cartridge.
Multiplex PCR
Determination of drug sensitivity to rifampin was diagnosed by designed tetra-arms pair primers targeted segments of rpoB gene mutation, rpoB 516 218 bps, rpoB 526 185 bps, rpoB 531 170 bps and RIRm respectively, 5´CAGCTGAGCCAATTCATGGA3´, 5 ´CTGTCGGGGTTGACCCA3´, 5 ´CACAAGCGCCGACTGTC3´ and 5 ´TTGACCCGCGCGTACAC3´. The amplification of all PCR reactions was performed in 200 μl micro-tubes and cycled in the program temp control system PC-320 thermo cyclers (ASTEC). The micro-tubes contained 25 μl PCR reaction consisted of 5 μl of isolated DNA, 5.5 mM of MgCl2, 0.2 mM of each dNTP, 1.25 hot start taq enzyme (iu) and 10 pmol/μl of each forward and reverse primers. Thermo cycler condition was as follows: Denaturation for 5 min at 95℃ and 40 cycles with 30 second at 95℃, 30 seconds at annealing temperature 68℃ and 72℃ for 30 seconds followed by the extension step at 72℃ for 7 min. Only 7 μl of the amplified products were loaded on 8% acrylamide gel. The DNA bands were visualized by a gel doc viber trans illuminator [3].
Real-time PCR
A 81-bp region covering the -bp “rifampin resistance-determining region” of the rpoB gene was targeted; primers rpoB F 5′- ACCGCAGACGTTGATCAACAT-3′, rpoB R 5′- GGCACGCTCACGTGACAG-3′, probes rpoB w 516 5′- FAMCATGGACCAGAACAACCCGCTGT– 3/-3′, rpoB w 526 5′- FAM-TCGGGGTTGACCCACAAGCG–3/-TAMRA -3′, rpoB w 531 5′- ROX-AGCGCCGACAGTCGGCG-BHQ-2 -3′. A total of 25 μl of PCR mixture was prepared. It included 0.2 Mm dNTP, 5.5 Mm Mgcl2, 10X Buffer, 10 Pmol rpoB F, 10 Pmol rpoB R, 5 Pmol Probes, Qbufer, 1U enzyme, 5-20 ngr DNA. The real-time PCR was performed in capillary tubes in the lightcycler instrument (Roche diagnostics). The tow step cycling conditions were 5 min at 95°C, 15 seconds at 95°C, 60 seconds at 61°C (with a single acquisition of fluorescence) in a total cycle 45-55. The fluorescence of the fluorescein was visualized through channel FAM, ROX and PET. In a sample sensitive to rifampin, CT (Cycle Thresholds) is less than or equal to 40 and in resistant samples CT is greater than 40, which are lower than the threshold line.
Restriction fragment length polymorphism PCR
RFLP was used to diagnose rpoB 531, rpoB 526 and rpoB 516 point mutations. For determination of rpoB 531, designed pair primers ROF; 5′-GTCGCCGCGATCAAGGA-3′, RIR 5′- TGACCCGCGCGTACAC-3′ and R531: 5′- ACAAGCGCCGACTGTC-3′ was used. PCR mixture for 25 μl total reaction was included of 0.5 μl dNTP, 1 μl Mgcl2, 0.15 μl enzyme (Taq), 2.5 μl PCR Buffer, 0.4 μl of each primer, 2 μl of template PCR product. The mixture was cycled for 4 min at 94° C as initial denaturation and then followed 30 cycles including denaturation 30 sec at 94°C, annealing 30 sec at 57°C and 1 min at 72°C followed with final extension 7 min 72°C. For detecting rpoB 526, 1 pmol ROF, 20 pmol RIR and 30 pmol R526 with sequence of ROF: 5′- GTCGCCGCGATCAAGGA-3′, RIR: 5′- TGACCCGCGCGTACAC-3′ and R526: 5′-GTCGG GGTTGACCCA-3′ was used. PCR mixture for 25 μl reaction was contained of 2.5 μl PCR buffer, 0.15 μl Enzyme (Taq), 1 μl Mgcl2 and 0.5 μl dNTP. The reaction was then cycled 4 min at 94°C as initial denaturation, 30 cycles of 30 sec at 94°C, 30 sec at 58.8 V and 1 min at 72°C with the final extension step 7 min at 72°C. For rpoB 516 point mutations, 1 pmol ROF 5′- GTCGCCGCGATCAAGGA-3′, 10 pmol 5′- TGACCCGCGCGTACAC-3′, 15 pmol 5′- GCTGAGCCAATTCATGGA-3′ was used. The 25 μl mixture was also included of 0.5 μl dNTP, 1 μl Mgcl2, 0.15 μl Enzyme (Taq) and 2.5 μl PCR buffer. Then the mixture was cycled for the initial denaturation for 4 min at 94°C. It was totally cycled at 30 included of 30 sec at 94°C annealing 30 sec at 53.7°C and 1 min at 72°C with a final extension 7 min 72°C. The amplified fragments were electrophoresed in 1.5% agarose gels. A single band at 248 bp indicated a mutant allele and wild-type isolates reported once two bands at 214 bp and 248 bp visualized.
Statistical analysis
This cross-sectional study has evaluated sensitivity and specificity, positive and negative predictive values of GeneXpert MTB/RIF compared to smear microscopy, multiplex PCR, RFLP-PCR and real time PCR was evaluated by SPSS13.0 software with 95% Confidence Interval (CI).
In the current study, a total of 93 patient samples were assessed. The mean age of patients was 62.75 years with a female predominance accounted for 59.1%. The pulmonary sample was 93 sputum. All 93 samples tested by GeneXpert MTB/RIF, of 73 (78.49%) positive samples, 59 (63.44%) and 17 (18.27%) were positively reported as susceptible and resistance cases respectively and also 17 (18.27%) were found to be negative. The positivity rates for microscopy, GeneXpert MTB/RIF, real time PCR, RFLP-PCR and multiplex PCR were 77.41%, 78.49%, 78.49%, 75.26% and 67.74%, respectively. Among all the GeneXpert results, only 3 cases were found as false negatives and no case has reported as false positive compared to RFLP-PCR. Comparing RFLP-PCR with GeneXpert results as the gold criterion, sensitivity and specificity were found 96% and 100%, respectively and it has reported as the most potent technique compared to others for diagnosis of Mtb and the genome sequence of rifampicin resistance. The sensitivity, specificity, PPV and NPV of all other methods compared with GeneXpert results, as the reference technique, are summarized in Table 1. Multiplex and RFLP PCR were found as the other potent diagnostic TB-resistant methods (Figures 1-4).
| Sample | Microscopic examination | Real-time PCR | RFLP-PCR | Multiplex PCR | ||||||||||||
| Sensitivity% | Specificity% | PPV% | NPV% | Sensitivity% | Specificity% | PPV% | NPV% | Sensitivity% | Specificity% | PPV% | NPV% | Sensitivity% | Specificity% | PPV% | NPV% | |
| Sputum 93 | 96 | 62 | 87 | 76 | 93.3 | 81.2 | 84 | 72 | 96 | 100 | 80 | 85 | 82.8 | 100 | 78 | 56 |
Table 1: The sensitivity, specificity, PPV and NPV of diagnostic methods compared to GeneXpert as the gold criterion.
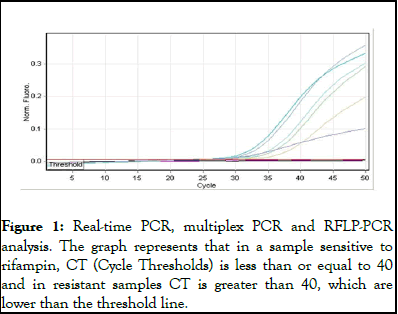
Figure 1: Real-time PCR, multiplex PCR and RFLP-PCR analysis. The graph represents that in a sample sensitive to rifampin, CT (Cycle Thresholds) is less than or equal to 40 and in resistant samples CT is greater than 40, which are lower than the threshold line.
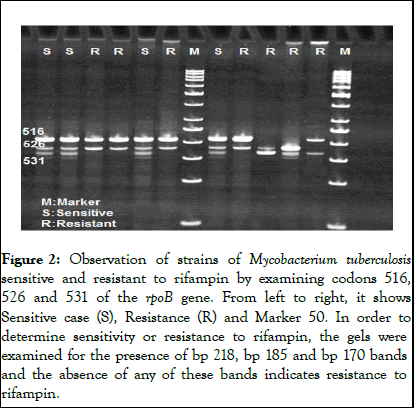
Figure 2: Observation of strains of Mycobacterium tuberculosis sensitive and resistant to rifampin by examining codons 516, 526 and 531 of the rpoB gene. From left to right, it shows Sensitive case (S), Resistance (R) and Marker 50. In order to determine sensitivity or resistance to rifampin, the gels were examined for the presence of bp 218, bp 185 and bp 170 bands and the absence of any of these bands indicates resistance to rifampin.
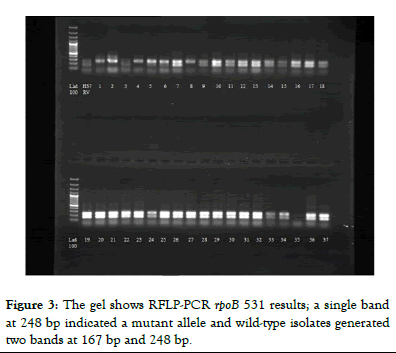
Figure 3: The gel shows RFLP-PCR rpoB 531 results; a single band at 248 bp indicated a mutant allele and wild-type isolates generated two bands at 167 bp and 248 bp.
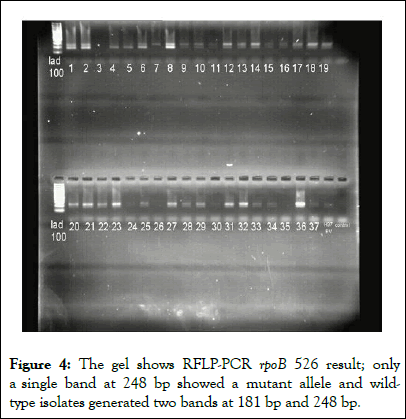
Figure 4: The gel shows RFLP-PCR rpoB 526 result; only a single band at 248 bp showed a mutant allele and wildtype isolates generated two bands at 181 bp and 248 bp.
Limited resources communities still face with tuberculosis as a public health threat with an increasing death rate. So, rapid diagnosis and planning proper treatment is ultimately important to diminish the mortality. Although culture is the gold standard technique, it is time-consuming, requires equipped laboratory and lab technical expertise [4]. The AFB smear is rapid and inexpensive, but, it is a fairly insensitive method (20%-80%) and due to limited specificity it cannot make a differentiation between Mtb and Non-Tuberculous Mycobacteria (NTM) strains. Other molecular amplification methods may also have varied sensitivity and specificity. The GeneXpert MTB/RIF as a diagnostic method is a new, rapid, easy-to-perform method that enables monitoring of the amplification and the detection of mutations. The GeneXpert uses DNA PCR amplification for both diagnosis of Mtb and rifampicin resistance- related mutations in a same time [5].
The present study enrolled a total of 93 pulmonary samples suspected of tuberculosis infection, from those admitted to the Massih Daneshvari hospital. We aim to compare the diagnostic accuracy of GeneXpert MTB/RIF diagnostic yield method as a reference standard with the AFB smear microscopy and some of molecular based diagnostic techniques, multiplex PCR, Restriction Fragment Length Polymorphism-PCR (RFLP-PCR) and real-time PCR. In this study, it was possible to detect 93.3% of the rifampin-resistant isolates by real-time PCR with two probes covering the regions from codons 531 to 516 and codons 526. Other mutations that are not covered by the probes used in this study may be responsible for rifampin resistance in the isolates that are not detected by real-time PCR. Its specificity, PPV and NPV were found 81.2%, 84% and 72% which documented this technique as a powerful diagnostic platform of MTB-RIF. One study by Tanil Kocagoz in 2005 reported the use of diagnostic real-time PCR for determination of rifampin resistance in clinical isolates of Mycobacterium tuberculosis. The results obtained by real-time PCR were compared to those obtained by the proportion method. For detection of rifampin resistance, the real-time PCR assay yielded a sensitivity of 92.7% and a specificity of 100%. In 2010, Catharina C. Boehme also evaluated molecular detection of tuberculosis and rifampin resistance. As compared with phenotypic drug-susceptibility testing, MTB/RIF testing correctly identified 200 of 205 patients (97.6%) with rifampin-resistant bacteria and 504 of 514 (98.1%) with rifampin-sensitive bacteria [6].
According to the current results, the sensitivity and specificity of AFB smear microscopy were lower than those of GeneXpert MTB/RIF assay 96% and 62% respectively. Out of the 93 samples, only 77.4% were detected by AFB smear. PPV and NPV were also reported 87% and 76% respectively. Due to different geographical regions and different laboratories settings, the sensitivity of AFB is varied and fairly insensitive. Smear microscopy staining is still considered for the reports to explore the degree of patients’ infectivity; the tuberculosis infectious dose is lower than ten bacilli. Meanwhile, the lower detection limit of AFB microscopy ranges from 5,000 to 10,000 AFB/ml; this means that AFB smear would miss many potentially infectious cases. Comparing smear microscopy with GeneXpert MTB/RIF assay in PTB detection, smear microscopy detected 72/93 MTB cases while GeneXpert assay also detected 72 including all true-positive cases of smear microscopy plus 6 positive cases among subjects with smear-negative results [7].
Muia et al., reported a sensitivity of 81.8% and specificity of 84.3% for smear microscopy compared to culture as the golden standard. Poor results were documented from other laboratories that showed sensitivity ranged from 20 to 80% and specificity of 74.5%-80.7%. The significant differences in sensitivity of AFB microscopy are related to various reasons such as: Sample preparation; slide examination; use of fluorescent versus conventional stains and a long term history of using antibiotics. In the current study, the false-negative rate of smear microscopy reached 0.03%. Similarly, Meawed and Shaker in 2016 reported that, the false-negative results of smear microscopy can be mainly due to incompetency to discriminate between drug-susceptible and drug-resistant strains of MTB owing to poor sample preparation which need experts. Meanwhile, culture being the gold standard, it proceeds for weeks up to months to yield results and depends on sophisticated laboratory facilities and skilled technicians [8].
The multiplex PCR assay was also used for detection of RIF resistance region. Based on the current results, this study has reached sensitivity, specificity, PPV and NPV for 82.8%, 100%, 78% and 56%, respectively. In 2019, Saira Salim evaluated a multiplex PCR for rapid diagnosis of drug resistant Mtb. For identifying drug resistance, the specificity and sensitivity of multiplex PCR in isolates was reported 100% and 100% for rifampicin, 100% and 71% for isoniazid and 100% and 60% for ethambutol, respectively. When compared to phenotypically resistance results, the Positive Predictive Value (PPV) was 100% each and the Negative Predictive Value (NPV) was calculated to be 100%, 74% and 71% for RIF, INH and EMB, respectively. Saira Salim in 2021 also has published a comparison between the efficacy of GeneXpert Mycobacterium tuberculosis-rifampicin with multiplex polymerase chain reaction for diagnosis of Mycobacterium tuberculosis and rifampicin resistance. Gene Xpert Mycobacterium tuberculosis-rifampicin assay detected all 84 (100%) rifampicin-resistant samples, while multiplex polymerase chain reaction detected 44 (52.3%) such samples. The values of sensitivity, specificity, positive predictive and negative predictive of Gene Xpert was respectively found 100%, but those values for multiplex polymerase chain reaction were 52%, 100%, 100% and 72% respectively [9].
PCR-Restriction Fragment Length Polymorphism (PCR-RFLP) is a simple and robust method for the rapid identification of RIFresistant Mtb. According to the results, the RFLP sensitivity and specificity compared to GeneXpert were found 96% and 100% respectively. PPV and NPV were also accounted 80% and 85% respectively. In 2016, one study from Pakistan by Muhammad Riaz reported drug resistant strains of Mycobacterium tuberculosis identified using PCR-RFLP. They have found identifying genetic mutations in drug-resistant strains through PCR-RFLP for isoniazid, ethambutol, streptomycin and ofloxacin. Ofloxacin resistance was found same as the resistance in isoniazid, ethambutol and streptomycin of 6.5% cases. Resistance to isoniazid was detected in 61% cases, 50.4% in ethambutol and 43.1% in streptomycin cases. It was concluded that PCR-RFLP is a powerful laboratory for the rapid identification of genetic variation in drug-resistant tuberculosis patients and may be routinely used to diagnose drug resistance at the earliest diagnosis [10].
The higher cost of the GeneXpert assay must be compared to the insensitive and poor specificity of AFB microscopy and even evaluated its advantages other than other molecular methods. Lack of Mycobacteria Growth Indicator Tube (MGIT) as a rapid liquid culture methods and the potential impact on the study, evaluating the diagnostic precision of the GeneXpert assay on only sputum as a sample tested in this study and finally shortage of studying the impact of the assay on patient’s outcomes; are the most limitations of the current study.
All participants provided written informed consent to participate in the study. Also, informed consent to publish data of the current study was obtained from all participants. All the techniques and methods were performed according to relevant guidelines and regulations. Also, the methods of the current study were approved by an institutional ethical board from Masih Daneshvari hospital, Tehran, Iran.
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
The authors declare that there are no competing interests.
Not applicable.
Citation: Farnia P, Javadi A, Seif S, Farnia P, Ghanavi J (2025) Comparison of Real-Time PCR, RFLP-PCR and Multiplex PCR with GeneXpert MTB/RIF for Diagnostic Rifampicin Resistance among Pulmonary Tuberculosis. Mycobact Dis. 15:538.
Received: 24-Apr-2024, Manuscript No. MDTL-24-30935; Editor assigned: 29-Apr-2025, Pre QC No. MDTL-24-30935 (PQ); Reviewed: 13-May-2024, QC No. MDTL-24-30935; Revised: 13-Jan-2025, Manuscript No. MDTL-24-30935 (R); Published: 20-Mar-2025 , DOI: 10.35248/2161-1068.25.15.538
Copyright: © 2025 Farnia P, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.