
Journal of Clinical Trials
Open Access
ISSN: 2167-0870

ISSN: 2167-0870
Research Article - (2022)Volume 12, Issue 3
Background: COAD is among the most prevalent malignancy, with a very high incidence rate. Crosstalk between cancer and interstitial cells significantly affects cancer development, modulated partly by chemokines production. When present in the tumor microenvironment, CXC chemokines have been shown to regulate tumor cell activity and influence immune cell transport, resulting in anti-tumor immune mechanisms and influencing the outcomes of the patient; nonetheless, the CXC chemokines expression levels in COAD, as well as their prognostic significance, have not yet been established.
Methods: This study used UALCAN, GeneMANIA, STRING, TRRUST, cBioPortal, TIMER, and GEPIA.
Results: The expression of CXC1/2/3/5/6/11/12/13/14/16/17 in COAD patients was shown to be significantly correlated with the pathological stage. A considerably improved prognosis was observed in patients with low transcriptional levels of CXCL9/10/11. Differentially expressed CXC chemokines exert roles that are predominantly correlated with the chemokine signaling pathway and interactions of cytokine–cytokine receptors. Our findings indicated that the transcriptional factors, including SP1, RELA, and NFKB1 are essential for the production of CXC chemokines. Furthermore, we discovered a substantial association between the CXC chemokines production and infiltration of 6 kinds of immune cells (CD8+ T cells, dendritic cells, B cells, CD4+ T cells, neutrophils, and macrophages,).
Conclusion: These findings might be useful in identifying prognostic indicators and immunotherapeutic targets for colon cancer.
Colon adenocarcinoma; Chemokines; Tumor immunity
Colon Adenocarcinoma (COAD) is a very prevalent kind of tumor. Moreover, it's the second most common kind of cancer in women and the third most prevalent type in males [1]. COAD is the most frequent histologic subtype of CRC, accounting for 90 percent of all CRCs. Despite technological advancements, COAD patients continue to experience an unfavorable prognosis, particularly in the case of metastases to distant organs or lymph nodes. Thus, comprehending the fundamental processes of COAD is critical for developing fresh approaches for enhanced COAD therapy [2,3]. Currently, several researchers have investigated the pharmacological targets of COAD, including immune checkpoint inhibitors and kinase. Nevertheless, these interventions are not adequate, and further therapeutic targets, as well as prognostic indicators, are of great necessity. Chemokines, a family of around 50 chemotactic cytokines with low molecular weight, are involved in a variety of biological processes, such as tumor progression, angiogenesis and metastasis, and leukocyte migration [4]. In the Tumor Microenvironment (TME), chemokines are released by tumor cells as well as by other cell types, such as stromal and immune cells. Chemokines have the ability to affect immune cell trafficking as well as lymphoid tissue formation, enabling them to regulate anti-tumor immune responses spatiotemporally [5]. Some data suggest that chemokines may influence tumor immunity as well as tumor immune and biological phenotypes both directly and indirectly [6]. In this way, carcinogenesis, progression, treatment impact, and patient outcomes are all influenced [7]. CXC chemokines are critical constituents of the chemokine family; they may serve as prognostic biomarkers and COAD treatment targets [8]. Numerous research reports have shown the overall expression and role of CXC chemokines in COAD, however, finding viable CXC chemokines that may serve as prognostic markers and treatment targets for COAD is challenging [9]. In view of the fast growth of multiple databases, it is now feasible to perform an extensive investigation of CXC chemokines. Hence, a detailed bioinformatics investigation of the CXC chemokines expression in COAD was performed in the present research. The potential of these biomarkers as therapeutic targets and prognostic indicators was assessed using various powerful public databases for the purpose of offering doctors relevant treatment medications for patients suffering from COAD.
GEPIA
We used GEPIA (http://gepia.cancer-pku.cn/index.html) to perform differential mRNA analysis on tumor and normal tissues [10], “Single Gene Analysis” module analysis in the pathological stage, and determine prognostics of CXC chemokines. The "Multiple Gene Comparison" module study of CXC chemokines was carried out utilizing the "COAD" dataset. The P-value threshold was defined as <0.05. The p-value for the expression or pathological stage analysis was determined utilizing the Student's t-test. For the purpose of performing a prognosis analysis, a Kaplan-Meier curve was employed.
UALCAN
Based on data from The Cancer Genome Atlas (TCGA) and the MET500 cohort, we utilized UALCAN (http://ualcan.path.uab.edu/ analysis.html) to carry out this procedure [11]. Using the “Expression Analysis” module and the “COAD” dataset, we obtained expression data of CXC chemokines. For the purpose of calculating a p-value, we employed the Student's t-test, with a threshold value of 0.05.
cBioPortal
For the purpose of evaluating multidimensional cancer genomics data, we utilized the cBioPortal (www.cbioportal.org) [12] and obtained CXC chemokines genetic alterations and co-expression.
GeneMANIA
Protein and genetic relationships, pathways, and protein domain similarities were discovered using GeneMANIA (http://www. genemania.org) [13].
STRING
We used STRING (https://string-db.org/) to integrate public sources of Protein-Protein Interaction (PPI) data [14] to analyze the differently expressed CXC chemokines and investigate their interactions.
TRRUST
We used TRRUST (https://www.grnpedia.org/trrust/) to obtain information on the regulation of Transcription Factor (TF)-target interactions [15].
TIMER
We used TIMER (https://cistrome.shinyapps.io/timer/) to obtain systematic evaluations of the infiltration status of immune cells and their clinical impact [16]. The "Gene module" was employed to investigate the relationship between immune cell infiltration and CXC chemokine levels. We utilized the "Survival module" to assess the clinical results of the expression of CXC chemokine and the infiltration status of immune cells.
RESULTS
CXC chemokine expression in COAD patients
We used the TCGA database to obtain sixteen CXC chemokines (excluding CXCL15). Using UALCAN, we determined the CXC chemokines expression levels in both normal tissues and the COAD tissues. The CXCL1 expression levels (p<1e−12), CXCL2 (p=1.62e−12), CXCL3 (p<1e−12), CXCL5 (p=1.05e−07), CXCL6 (p=5.27e−11), CXCL11 (p=4.20e−11), CXCL16 (p<1e−12), and CXCL17 (p=3.83e−05) in COAD tissues were considerably elevated, whereas the levels of transcription of CXCL12 (p=2.02e−10), CXCL13 (p=1.44e−02), and CXCL14 (p=1.61e−02) were significantly attenuated. These results are presented in Figure 1.
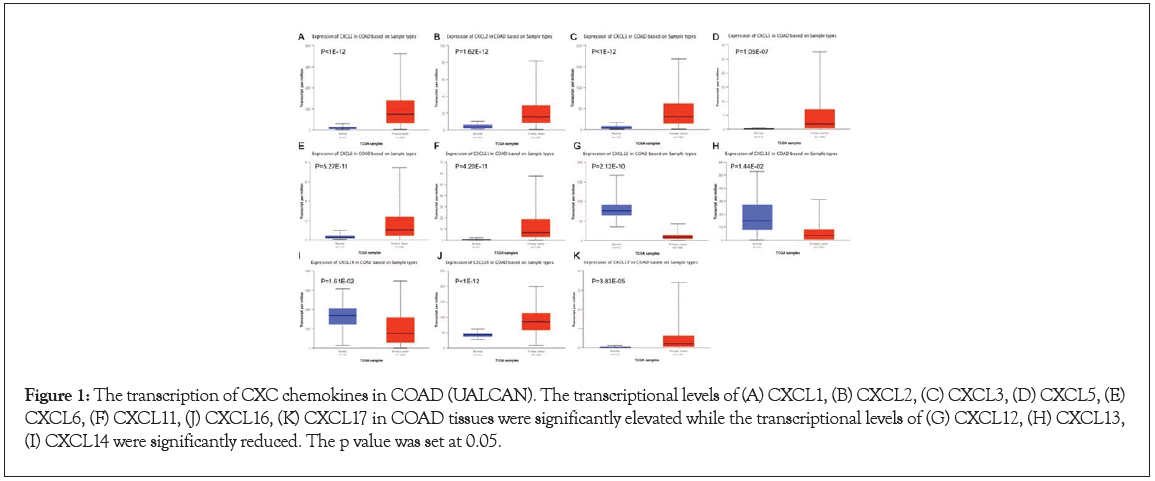
Figure 1: The transcription of CXC chemokines in COAD (UALCAN). The transcriptional levels of (A) CXCL1, (B) CXCL2, (C) CXCL3, (D) CXCL5, (E) CXCL6, (F) CXCL11, (J) CXCL16, (K) CXCL17 in COAD tissues were significantly elevated while the transcriptional levels of (G) CXCL12, (H) CXCL13, (I) CXCL14 were significantly reduced. The p value was set at 0.05.
We also analyzed the relative expression levels regarding CXC chemokines in COAD tissues and discovered that CXCL16 had the highest relative expression level of any of the CXC chemokines we assessed (Figure 2). For the purpose of finding other CXC chemokines that are correlated with the progression, carcinogenesis, as well as clinical outcomes of COAD patients, we analyzed all of the CXC cytokines shown to be differently expressed between COAD tumors and normal tissues (CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL11, CXCL12, CXCL13, CXCL14, CXCL16, and CXCL17). CXCL4, CXCL7, CXCL8, CXCL9, CXCL10, and CXCL15 were inapplicable in the subsequent investigation because their expression levels were similar both in COAD tumors and normal tissues. We investigated the relationship between the differentially expressed CXC chemokines and the clinical stage of COAD patients, where we observed a considerable correlation between the pathological stage and the CXCL9 (p=0.003), CXCL10 (p=0.006), and also CXCL11 (p=0.004) (Figure 3). The expression levels of CXCL9, CXCL10, and CXCL11 were elevated with the progression of the tumor. According to the findings, these CXCL chemokines perform a critical role in the occurrence and progression of COAD tumors.
Figure 2: The relative levels of CXC chemokines in COAD.
Figure 3: Correlation between different expressed CXC chemokines and the pathological stage of COAD patients (GEPIA). p<0.05. (A)(N) CXCL1CXCL14 and (O)(P) CXCL16CXCL17.
Prognostic significance of CXC chemokines in COAD patients
We used GEPIA to evaluate CXC chemokine expression in COAD progression, comparing the associated expression of CXC chemokines with clinical outcomes. Figure 4 depicts disease-free survival curves. The presence of high transcriptional levels of CXCL9 (p=0.026), CXCL10 (p=0.0011), and CXCL11 (p=0.0045) in COAD patients was shown to be correlated with a prolonged Disease-Free Survival (DFS) time. The expression of the CXC chemokine was also examined in relation to the survival of COAD patients. We discovered that COAD patients with high transcriptional levels of CXCL2 (p=0.042), CXCL3 (p=0.047), and CXCL14 (p=0.045) had substantially longer overall lifespan (Figure 5).
Figure 4: The prognostic value of different expressed CXC chemokines in COAD patients in the disease free survival curve (GEPIA). The disease free survival curve of (A)(N) CXCL1CXCL14 and (O)(P) CXCL16CXCL17 in COAD.
Figure 5: The prognostic value CXC chemokines in COAD patients in the overall survival curve (GEPIA). The overall survival curve of (A)(N) CXCL1CXCL14 and (O)(P) CXCL16CXCL17 in COAD.
CXC chemokine gene mutation, interaction, and coexpression analysis in patients with COAD
We carried out a thorough investigation of the molecular features of the CXC chemokines. Genetic changes were investigated using the TCGA database. We found that CXCL1, CXCL2, CXCL3, CXCL8, CXCL9, CXCL10, and CXCL17 were altered in 5, 5, 5, 5, 5, 5, and 5% of the retrieved COAD samples (Figure 6A). There is a potential co-expression among the CXC chemokines that are differently expressed. We used STRING to create a PPI network to examine CXC chemokines with differential expression for the purpose of investigating the possible connections between them. In the PPI network, we discovered multiple nodes with 8 and several edges with a 43 that were acquired from it (Figure 6B).
Figure 6: Genetic alteration, neighbor gene network, and interaction analyses of different expressed CXC chemokines in COAD patients. (A) Summary of alterations in different expressed CXC chemokines in COAD. (B,C) Protein-protein interaction network of different expressed CXC chemokines.
The inflammatory responses, as well as the chemokine signaling pathway, were shown to be correlated with the role of CXC chemokines. The findings of GeneMANIA also indicated that the roles of CXC chemokines with differential expression were mostly correlated with chemokine activities, cell chemotaxis, as well as chemokine receptor binding, (Figure 6C).
Transcription factor target COAD patients
In view of the considerable difference in the CXC chemokines expression between COAD tissues and normal tissues, we investigated the possibility that differentially expressed CXC chemokines had transcription factor and kinase targets utilizing TRRUST databases including CXCL1, CXCL2, CXCL5, CXCL8, CXCL10, CXCL12, as well as CXCL14. Three transcription factors (NFKB1, RELA, and SP1) were discovered to be correlated with the modulation of CXC chemokines (Table 1). The hub transcriptional factors for CXCL1, CXCL2, CXCL5, CXCL8, CXCL10, and CXCL12 were RELA and NFKB1. Furthermore, SP1 was the primary transcriptional factor involved in the expression of CXCL1, CXCL5, and CXCL14. CXC Chemokines Immune Cell Infiltration in COAD Patients COAD patients have been demonstrated to have altered clinical outcomes after being exposed to CXC chemokines, which are correlated with immune cell infiltration and inflammatory reactions. In view of this, we utilized the TIMER algorithm to examine the relationship between immune cell infiltration and differentially expressed CXC chemokines. The findings illustrated a negative association between macrophages infiltration (Cor=−0.15, p=2.46e−03) and CXCL1 expression, and a positive association between CXCL1 expression and neutrophils infiltration (Cor=0.364, p=4.93e−14), CD8+ T cells infiltration (Cor=0.122, p=1.36e−02), and dendritic cells infiltration (Cor=0.177, p=3.72e−04) (Figure 7A).
| Key TF | Description | List of overlapped genes | p value | Q value |
|---|---|---|---|---|
| RELA | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | CXCL10,CXCL8,CXCL12,CXCL2,CX CL5,CXCL1 | 4.22E-08 | 6.58E-08 |
| NFKB1 | nuclear factor of kappa light polypeptide gene enhancer in Bcells 1 | CXCL12,CXCL8,CXCL10,CXCL5,CX CL1,CXCL2 | 4.39E-08 | 6.58E-08 |
| SP1 | Sp1 transcription factor | CXCL5,CXCL1,CXCL14 | 0.00461 | 0.00461 |
Table 1: Key regulated factor of CXC chemokines in RCC (TRRUST).
Figure 7: The Correlation between different expressed CXC chemokines and immune cell interaction (TIMER). The Correlation between the abundance of immune cell and the expression of (A) CXCL1, (B) CXCL2, (C) CXCL3, (D) CXCL5, (E) CXCL6, (F) CXCL9, (G) CXCL10, (H) CXCL11, (I) CXCL12, (J) CXCL13, (K) CXCL14, (L) CXCL16, (M) CXCL17 in COAD.
CXCL2 expression was correlated with the macrophages infiltration (Cor=−0.197, p=6.80e−05) in a negative manner, and correlated with the neutrophils infiltration (Cor=0.265, p=7.14e−08) (Figure 7B) in a positive manner. CXCL3 expression was shown to be negatively correlated with the infiltration of macrophages (Cor=−0.229, p=3.48e−06), and positively correlated with the infiltration of neutrophils (Cor=0.257, p=1.86e−07) (Figure 7C). CXCL5 expression was shown to exhibit a positive correlation with the macrophages infiltraton (Cor=0.148, p=2.88e−03), CD8+ T cells infiltration (Cor=0.151, p=2.27e−03), neutrophils infiltration (Cor=0.435, p=5.46e−20), as well as dendritic cells infiltration (Cor=0.22, p=8.14e−06) (Figure 7D). Furthermore, CXCL6 expression was found to be positively correlated with the macrophages infiltration (Cor=0.218, p=9.80e−06), CD8+ T cells infiltration (Cor=0.226, p=4.07e−06), neutrophils infiltration (Cor=0.456, p=6.06e−22), and also dendritic cells infiltration (Cor=0.277, p=1.67e−08) (Figure 7E).
CXCL9 expression was illustrated to positively correlated with the CD8+ T cells infiltration (Cor=0.488, p=1.25e−25), B cells infiltration (Cor=0.267, p=4.99e−08), CD4+ T cells infiltration (Cor=0.292, p=2.45e−09), neutrophils infiltration (Cor=0.684, p=1.27e−56), dendritic cells infiltration (Cor=0.632, p=3.63e−46) and macrophages infiltration (Cor=0.428, p=1.81e−19) (Figure 7F).
The same outcomes were acquired for CXCL10, CXCL11, CXCL12, CXCL13, CXCL14, CXCL16, and CXCL17. CXCL10, CXCL11, CXCL12, CXCL13, and CXCL16 expression was shown to be positively correlated with the infiltration of CD8+ T cells, B cells, CD4+ T cells, neutrophils, dendritic cells, as well as macrophages (all p<0.05) (Figures 7G–7L). Expression of CXCL14 was negatively correlated with neutrophils infiltration (Cor=-0.163, p=1.09e−03), CD8+ T cells infiltration (Cor=-0.141, p=4.33e−03), and dendritic cells infiltration (Cor=-0.109, p=2.91e−02), and showed positive correlation withthe infiltration of CD4+ T cells (Cor=0.186, p=1.80e−04) (Figure 7K). Expression of CXCL17 was positively correlated with B cell infiltration (Cor=0.201, p=4.59e−05) (Figure 7M).
The relationship between CXC chemokines with differential expression and the infiltration status of immune cells was investigated. Specifically, the Cox proportional hazards model was employed in the present research, which was modified to take into account the confounders below: dendritic cells, CD8+ T cells, macrophages, B cells, CD4+ T cells, neutrophils, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL9, CXCL10, CXCL11, CXCL12, CXCL13, CXCL14, CXCL16, and CXCL17. The expression of CXCL13 (p=0.020) and CXCL14 (p=0.012) was shown to have a substantial correlation with the clinical outcomes of COAD patients (Table 2).
| coef | HR | 95%CI_l | 95%CI_u | p value | |
|---|---|---|---|---|---|
| B_cell | 2.009 | 7.453 | 0.055 | 1016.962 | 0.423 |
| CD8_Tcell | -2.907 | 0.055 | 0.001 | 4.251 | 0.191 |
| CD4_Tcell | 2.502 | 12.202 | 0.058 | 2558.281 | 0.359 |
| Macrophage | 1.263 | 3.535 | 0.023 | 550.261 | 0.624 |
| Neutrophil | -1.285 | 0.277 | 0 | 2541.136 | 0.783 |
| Dendritic | 2.103 | 8.191 | 0.35 | 191.632 | 0.191 |
| CXCL1 | -0.29 | 0.748 | 0.532 | 1.052 | 0.095 |
| CXCL2 | -0.043 | 0.958 | 0.69 | 1.33 | 0.796 |
| CXCL3 | 0.202 | 1.224 | 0.794 | 1.887 | 0.359 |
| CXCL5 | 0.018 | 1.018 | 0.873 | 1.187 | 0.822 |
| CXCL6 | 0.176 | 1.193 | 0.885 | 1.607 | 0.247 |
| CXCL9 | 0.13 | 1.139 | 0.821 | 1.581 | 0.436 |
| CXCL10 | -0.012 | 0.989 | 0.674 | 1.449 | 0.953 |
| CXCL11 | -0.129 | 0.879 | 0.693 | 1.115 | 0.289 |
| CXCL12 | 0.011 | 1.011 | 0.796 | 1.284 | 0.928 |
| CXCL13 | -0.291 | 0.748 | 0.585 | 0.955 | 0.02 |
| CXCL14 | -0.141 | 0.869 | 0.778 | 0.969 | 0.012 |
| CXCL16 | -0.035 | 0.966 | 0.689 | 1.354 | 0.84 |
| CXCL17 | 0.077 | 1.081 | 0.93 | 1.256 | 0.313 |
Table 2: The cox proportional hazard model of CXC chemokines and six tumor-infiltrating immune cells in COAD (TIMER).
CXC chemokines were originally inflammatory markers, performing a critical function in angiogenesis, progression, and metastasis of tumors, and leukocyte migratory activities. A prior investigation established that CXC chemokines have a key role in carcinogenesis, tumor cell proliferation, and death, as well as tumor metastasis [17]. In the present research, intercellular cross-talk between stromal and COAD cells was distinct, which influences the chemokines expression profiles, thus facilitating the formation of a specific microenvironment for metastasis and tumor invasion [18]. Despite the fact that the characteristics of tumor immunophenotypes have been defined [19], the cell specificity of CXC chemokines expression signals is largely unknown. The predictive power of CXC chemokines in COAD, as well as their biological mechanisms, has not been well investigated.
In the present research, the expression of CXC chemokines in COAD and their correlations with the clinical stage were investigated. When comparing COAD tissues to normal tissues, we discovered that 11 genes were differently expressed (upmodulation of CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL11, CXCL16, and CXCL17; down-modulation of CXCL12, CXCL13, and CXCL14). Furthermore, we discovered that the expression levels of CXC9, CXC10, and CXC11 were elevated with the progression of the malignancies. We discovered that differently expressed CXC chemokines might perform a crucial function in COAD and that CXCL16 expression was considerably elevated in COAD tissues. COAD patients with elevated CXCL9, CXCL10, and CXCL11 expression levels exhibited a considerably longer DFS time, while patients with elevated CXCL2, CXCL3, and CXCL14 expression levels had a considerably longer OS time. We investigated the molecular properties of various chemokines found in COAD. The CXC chemokines differently expressed in COAD had genetic variants. Tumorigenesis and the progression of COAD are complicated and multifaceted processes in which genetic variation performs a significant function in the process [20].
By means of STRING and GeneMANIA analysis, we examined the role of differently expressed CXC chemokines. As predicted, we discovered that the activities of these genes are correlated with the chemokine signaling pathway and the interactions of cytokines with their receptors. We also identified the kinase targets and transcription factors of differently expressed CXC chemokines and discovered that NFKB1, SP1, and RELA, might be the predominant transcriptional factors involved in CXC chemokines modulation. The role of RELA in mediating oncogene-induced senescence in preneoplastic lesions had been revealed in a previous report [21]. By inhibiting the aberrant signaling pathway, NFKB1, which reduces inflammation status and the progression of cancer, has been shown to perform an inhibitory effect in the occurrence and progression of malignancies [22].
Chemokines aid in the migration as well as the localization of the immune cells [23]. There is proof that immune cell infiltration may have an impact on tumor progression and recurrence, clinical outcomes, and immunotherapy [24]. We discovered a relationship between the expression levels of CXC chemokines and the infiltration of the six distinct types of immune cells, including CD8+ T cells, dendritic cells, B cells, macrophages, CD4+ T cells, and neutrophils, showing that CXC chemokines may also be used to assess the immunological state. In the present research, we investigated the expression levels as well as the prognostic significance of CXC chemokines in a variety of malignancies, although the present research was restricted in scope. Hence, further independent investigations are necessary in order to validate these findings.
In summary, the present research will bring novel clues that will assist doctors in selecting suitable medications and prognostic biomarkers, as well as in identifying biomarkers that can robustly anticipate the survival profiles of COAD patients. These findings might be useful in identifying prognostic indicators and immunotherapeutic targets for colon cancer.
Data availability
The authors confirm that all of the original data used in the present research can be acquired from publicly available resources. The TCGA database (https://tcga-data.nci.nih.gov/tcga/) was used to acquire both the transcriptome and clinical data for COAD. Other data that were utilized to support the conclusion of the present research may be found in the supplementary information files. Upon request, the first author or the corresponding author will provide access to all of the raw data used in the present research.
Any repository data used in this study are open access and do not require any permissions. Ethics approval and consent to participate are not applicable for them.
The authors declare that they have no competing interests.
The authors state that they have received no support from any sources that should be acknowledged.
We are grateful to the TCGA databases for making the information available.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Wang H, Guo C, Luo J, Li Q, Fubuqing B, Jiang W (2022) Biomarkers and Targets Identification among CXC Chemokines in the Colon Adenocarcinoma. J Clin Trials. 12:500.
Received: 13-May-2022, Manuscript No. JCTR-22-17571; Editor assigned: 16-May-2022, Pre QC No. JCTR-22-17571 (PQ); Reviewed: 30-May-2022, QC No. JCTR-22-17571; Revised: 06-Jun-2022, Manuscript No. JCTR-22-17571 (R); Published: 13-Jun-2022 , DOI: 10.35248/2167-0870.22.12.500
Copyright: © 2022 Wang H, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.