Journal of Probiotics & Health
Open Access
ISSN: 2329-8901
ISSN: 2329-8901
Research - (2019)Volume 7, Issue 2
Probiotics in various combinations and in various forms have become popular supplements for establishing and maintaining gut health. However, commercial products vary in the effectiveness of their specific probiotic strains and the ability of these strains to survive the acidic environment of the stomach. A novel probiotic, manufactured by BIOHM, LLC, has a unique formulation of Saccharomyces boulardii, Lactobacillus acidophilus, L. rhamnosus, and Bifidobacterium breve, in combination with amylase, which has been developed to re-balance the bacterial and fungal population of the human gastrointestinal tract and combat digestive biofilms formed by pathogenic bacteria and fungi. Our data shows that these strains have the ability to survive the acidic environment when taken within 30 minutes of meal, a factor that is vital to ensure probiotic effectiveness.
BIOHM; Acidic environment; Viability
Recent technological advances in DNA sequencing have enabled scientists to identify many of the vast numbers of microorganisms in the human gastrointestinal intestinal (GI) tract. Relatively new data have shown that not only bacteria but also fungi, and their interactions with each other, have a great influence on both the healthy (i.e. balanced) and diseased (dysbiotic) states of the human gut. Strains of bacteria and fungi in a healthy intestinal environment will benefit from each other ’ s metabolic by-products, enabling each to thrive. For example, Candida is able to break down the starch in carbohydrate rich foods, providing a source of simple sugars that are fermented by beneficial bacteria such as Prevotella and Ruminococcus [1]. On the other hand, dysbiosis, or an imbalance in the relative numbers of bacteria and fungi, may lead to a variety of disease states, including inflammatory bowel diseases [2-5]. In these cases, microbial interactions result in the formation of biofilms, also known as digestive plaque, which serve to protect the bacterial-fungal matrix from the effects of therapeutic agents such as antibiotics, as well as from the host immune system [6].
Certain probiotic bacteria have been evaluated for their potential to re-establish the microbial balance and thus prevent or reverse the adverse effects of such diseases as vaginal and oral yeast infections and GI tract infections [7-11]. In previous studies, we were able to identify probiotic species of bacteria and fungi that are able to support the beneficial GI microbes while simultaneously antagonizing pathogenic ones [12]. These microbes, which include Saccharomyces boulardii, Lactobacillus acidophilus, L. rhamnosus, and Bifidobacterium breve in combination with amylase, have been shown to prevent and treat biofilms in vitro [6].
These living probiotic strains, however, would be ineffective if they are not able to survive the trip through the digestive tract, which is known to have varying pH levels. The stomach, for instance, has a highly acidic environment (a low pH) especially when fasting [13,14]. Therefore, it is important to evaluate the ability of probiotic strains to survive in the different gastric acidic levels (acidic pH).
Probiotic strains
The four BIOHM probiotic strains included Saccharomyces boulardii ATCC MYA-796 strain SB 48, Lactobacillus rhamnosus ATCC 39595 strain LB 20, Lactobacillus acidophilus, ATCC 43121 strain RP 32, and Bifidobacterium breve ATCC 15701 strain S 46. Strains were retrieved from lyophilized stock according to the methods recommended by the American Type Culture Collection (ATCC) and preserved under -80°C freezer in Protect-Select vials. All strains were identified with sequencing of internal transcribed spacer (ITS) regions (yeast) and partial 16S regions of ribosomal DNA (bacteria) before use. From frozen cultures, lactobacilli were transferred to De Man, Rogosa and Sharpe agar (MRS), Bifidobacterium to Reinforced Clostridial agar (RCA), and S. boulardii to Sabouraud Dextrose (SD) agar plates and incubated at 37°C overnight (B. breve anaerobically). An inoculum of 5 × 105 was prepared using a hemacytometer or spectrophotometer for yeast and bacteria, respectively. Inocula (100 μl) were added to 5 ml of MRS or SD broth with the pH adjusted to 1.5, 2.5, and 3.5 with 1N hydrochloride solution. Inoculum in 5 ml of standard MRS broth (pH 5.7 to 6.58) and SD broth (pH 5.53) served as controls.
Tubes were incubated at 37°C, and at 0, 30, 60, and 120 minutes after inoculation, 100-μL aliquots of the cell suspension from each tube were serially diluted with MRS (both Lactobacillus species), RC (Bifidobacterium), or SD (S. boulardii) broth and spread on corresponding agar plates. Plating was performed in triplicate and incubated as described above for the bacteria and yeast. The numbers of colony forming units (CFUs) were determined after 24-48 hours of incubation.
Statistical analysis
The average log CFUs/mL ± the standard deviation (SD) was determined for each probiotic strain at each pH level over time was compared to the log CFUs at the 0 time point. The significance of differences were calculated by a one-way ANOVA using PRISM version 6.04 (Figure 1).
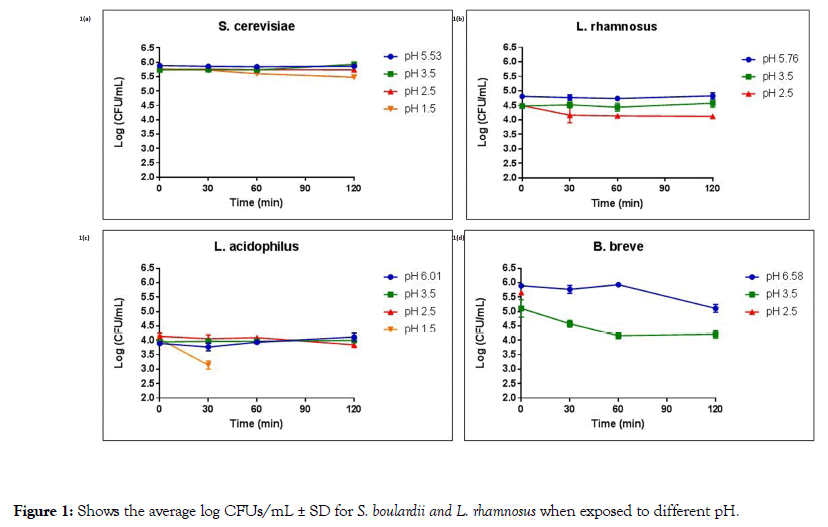
Figure 1. Shows the average log CFUs/mL ± SD for S. boulardii and L. rhamnosus when exposed to different pH.
Figure 1a shows the average log CFUs/mL ± SD for S. boulardii exposed to pH 5.53 (control), pH 3.5, pH 2.5, and pH 1.5 over a 120 minutes time span. As can be seen, S. boulardii demonstrated no significant difference in average log CFUs/mL for all pH levels and all time points tested, indicating that S. boulardii suffered no decrease in viability at pH ranges of 5.53-1.5 after 2 hours of exposure.
Figure 1b shows the average log CFUs/mL ± SD for L. rhamnosus exposed to pH 5.76 (control), pH 3.5, and pH 2.5 over a 120 minutes time span. L. rhamnosus demonstrated no significant difference in average log CFUs/mL for pH levels of 5.76-2.5 and all time points tested, indicating that L. rhamnosus also suffered no decrease in viability at pH ranges of 5.76-2.5 after 2 hours of exposure. However, L. rhamnosus showed no growth in pH 1.5 at all time points.
Figure 1c shows the average log CFU/mL ± SD for L. acidophilus exposed to pH 6.01 (control), pH 3.5, pH 2.5, and pH 1.5 over a 120 minutes time span. Our data showed that exposing L. acidophilus to pH 6.01 and 3.5 did not affect the viability of the organism. When incubated at a pH of 2.5 for 120 minutes L. acidophilus demonstrated a slight reduction in cell viability (p value=0.049), with 93.0% viability compared to the 0-time point. Moreover, at a pH of 1.5, L. acidophilus showed a significant decrease in average log CFUs/mL after 30 minutes exposure and did not survive after 60 minutes exposure (p values of <0.001).
Figure 1d shows the average log CFUs/mL ± SD for B. breve exposed to pH 6.58 (control), pH 3.5, and pH 2.5 over a 120 minutes time span. After 120 minutes in the control pH (6.58), B. breve demonstrated 86.7% viability, a significant decrease from the starting inoculum at the 0-ime point (p value of <0.001). Similarly, at a pH of 3.5, B. breve demonstrated significantly less growth at the 30, 60 and 120 minutes time points (89.3, 81.3 and 82.3% viability, respectively) when compared to the 0-time points (p values of 0.045, 0.002 and 0.001, respectively). No growth was observed for B. breve at a pH of 2.5.
Our data show that the S. boulardii and L. rhamnosus demonstrated viability when exposed to various pH levels ranging from 5.76-1.5 over a 2 hour period, while L. acidophilus showed viability at pH 6.01 and 3.5. At pH 2.5, L. acidophilus maintained its growth levels with no significant change in viability through 60 minutes and 93.0% viability after 120 minutes. B. breve at pH 6.58 showed sustained viability through 60 minutes, with a reduction to 86.7% at 120 minutes. Moreover, B. breve at a pH 3.5 showed ≥ 81.3% viability over a time span of 120 minutes.
Variable acidic environments influence multiple aspects of the functioning of the human GI tract. For example, it is important to determine the effect of pH levels on therapeutics and supplements administered by the oral route. Extensive studies have been performed to determine the effects of pH on the absorption of nutrients from foods [15], as well as on therapeutic drugs such as cancer drugs and antifungal agents [16,17]. Data from these studies have demonstrated that a highly acidic environment often has a negative effect upon absorption.
Furthermore, previous studies have been conducted to determine the role of low pH in the “ gastric bactericidal barrier”, which protects against certain pathogenic organisms. Giannella et al. reported that greater than 99% of Salmonella, E. coli, and Serratia marcescens strains were killed when exposed to gastric juices at pH 2.0-3.0 for as little as 15 minutes [18]. As such, the acidity of the stomach is an important protection against development of serious gastrointestinal diseases like cholera and dysentery [19,20].
Thus, it stands to reason that low pH may have deleterious effects on any probiotic strains that must survive passage through the stomach. Estimates of probiotic strain survival through the stomach range from ~20%-40% [21], a factor that manufacturers account for by overloading the initial organism load (referred to as overage) in order to insure delivery of adequate viable cells to the lower GI tract. Besides this overloading practice, one avenue that has been used to provide protection of beneficial probiotics is through microencapsulation. Krasaekoopt et al. showed that strains of Lactobacillus acidophilus and L. casei had greater survival rates in low pH when encapsulated into alginate beads coated with chitosan, though Bifidobacterium bifidum did not survive either as free cells or within encapsulated beads [22]. While Ding et al. reported similar negative results with microencapsulated Bifidobacterium, other studies have determined that microencapsulation is effective in preserving the viability of B. bifidum [23-25].
In light of conflicting reports on the effectiveness of physical means such as microencapsulation, it is important to determine which probiotic strains are not only effective in preventing or correcting dysbiosis in the gut but which are capable of surviving as free cells in the acidic environment of the stomach. Studies have shown that after a meal, the pH of the stomach can rise to a range of 4.0-6.0. Approximately 2 hours after eating, the pH will then return to pre-ingestion levels [26]. Our data show that S. boulardii and L. rhamnosus can survive while passing through the stomach at the fasting pH of 1.5. This is in agreement with the findings of Edwards-Ingram et al. where in S. boulardii survivability was significantly greater than S. cerevisiae, with S. boulardii survival rates similar to the control at pH 2 as opposed to only 4% survival rate for S. cerevisiae. Moreover, our study showed that both L. acidophilus and B. breve are able to survive in the stomach following a meal when the pH rises to between 3.5 and 6.0 [27].
Although these probiotic strains were tested as free cells, the commercialized BIOHM probiotic is enterically coated using hydroxymethyl cellulose known to provide protection against stomach acidity [28]. Furthermore, the overage in the initial microbial load more than compensates for the loss of viability (<20%) after exposure to an acidic environment reported above. Thus, not only has our BIOHM probiotic has been shown to deliver effective levels of probiotic strains when taken within 30 minutes of the completion of a meal, it provides the advantage of a beneficial yeast organism with greater acid resistance than some other commercial products.
Citation: Ghannoum M, Ghannoum A, Long L, Sun PL, Isham N (2019) BIOHM Probiotics Retain Viability In Low pH Environments Simulating The Digestive Environment. J Prob Health. 7:211. doi: 10.35248/2329-8901.19.7.211
Received: 24-Jul-2019 Accepted: 31-Jul-2019 Published: 07-Aug-2019 , DOI: 10.35248/2329-8901.19.7.211
Copyright: © 2019 Ghannoum M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.