Journal of Agricultural Science and Food Research
Open Access
ISSN: 2593-9173
ISSN: 2593-9173
Review Article - (2023)Volume 14, Issue 1
Molecular breeding in Ethiopia is in a very gradual introduction process. Beyond the detail understanding and application a technique or procedure factors, establishment cost is a paramount headache. Though, these factors limit its application in Ethiopia context different molecular breeding techniques such as TILLING and Eco TILLING are coming front especially at a university levels in theoretical basis. Targeting induced local lesions in genomes) is a broad reverse-genetic strategy for locating an allelic series of induced point mutations in genes of interest. TILLING allows for the rapid and cost-effective detection of induced point mutations in chemically mutagenized populations. The technique is applicable not only to model organisms but also to economically important plant organisms. TILLING provides a powerful approach for gene discovery, DNA polymorphism assessment, and plant improvement due to several advantages such as simple procedure, high sensitivity, and high efficiency. TILLING and EcoTILLING, when combined with other genomic resources, can be used immediately as a haplotyping tool in plant breeding for identifying allelic variation in genes exhibiting expression correlating with phenotypes and establishing. Thus, this review was examined in 2018 at Bahir Dar college of agriculture and environmental science to create understanding on modern crop improvement methods rather than conventional breeding in this era. Keywords: Eco TILLING; Induced mutation; Nucleotide sequence; TILLING
Eco TILLING; Induced mutation; Nucleotide sequence; TILLING
ATP: Arabidopsis TILLING Project; CEL I: Celery nuclease; DEB: Di Epoxy Butane; DHPLC: Denaturing High-Performance Liquid Chromatography; DNA: Deoxyribo Nucleic Acid; EMS: Ethyl Methyl Sulfonate; ENU: N-ethyl-N-nitroso Urea; PCR: Polymerase Chain Reaction; SNPs: Single Nucleotide Polymorphisms; STP: Single Plant Threshed; TILLING: Targeting Induced Local Lesions IN Genomes.
Variation in nucleotide sequence is mainly determine heritability of phenotypic differences and has been exploited by human for the improvement of crops since the starting of domestication. Variation either can be natural, from divergence populations, or induced through treatment application with mutagens [1,2].
As cited in, TILLING (Targeting Induced Local Lesions IN Genomes) is a non-transgenic reverse genetic technique that is suitable for most plants. For decades, mutations are created by treatment with the application of the same chemical mutagens have been employed for TILLING successfully.
TILLING could give mutations that are missense in allelic series and truncation through inducing primarily random point mutations at high density using chemical mutagens [3]. Whereas, EcoTIILING, an expansion of TILLING technique, can be used to discover point mutations or polymorphisms in natural populations [4]. Therefore, TIILING and EcoTIILING allow identifying diverse version of genes in germplasm and acquiring information about their gene function and provide guidelines so as to develop new strategies for genetic improvement of plants.
The difference between two techniques stated that, TILLING relies or depends on chemical methods to induce mutations in a given plant population and it uses various advanced technologies to find and use desirable gene versions or variants in the mutated population. EcoTIILING, on the other hand, relies on naturally occurring mutations rather than induced mutations, using advanced technologies similarly with TILLING to find and use desirable natural gene variants [5].
TILLING and Eco TILLING also can be identify unknown and known point mutations from a set of candidate genes both methodologies provide proof of function for both natural and induced variations. Reverse genetics techniques allow recognition of possible loss of functions for a particular gene during early stage of development [6]. Thus this review was go through to the application of TILLING and Eco TILLING in crop improvements
Discovery of TILLING
First, TILLING was developed in 2000 from the model plant Arabidopsis thaliana. This technique is successfully adapted and applied to several animal and plant species based on independent of their genome size, reproductive system, and generation time and ploidy level [7,8].
TILLING identifies induced mutations in mutagenized populations, the modified form known as Eco TILLING detects naturally occurring SNPs, especially in landraces and wild accessions [9]. The latter has additional applications in genetic mapping, breeding and genotyping, and also provides information concerning gene structure, linkage disequilibrium, population structure, or adaptation [10].
TILLING has proved to have additional benefits in addition to identifying polymorphism in genomes. The first part of TILLING requires the development of large number of mutagenized populations. Mutagenesis has been widely applied by breeders for many decades as a conventional improvement technique, and it has enabled the release of many crop cultivars [11-13]. However, through TILLING, this random mutagenesis is better exploited by screening for mutations in defined genes controlling the trait of interest. Since TILLING directly introduces genetic variation on improved or elite germplasm, it avoids the need for introgression of a mutant allele in a nonadapted background into current high-yielding varieties and avoids the problem of linkage drag. Therefore, the introduction of agriculturally undesirable traits is avoided [14-16].
TILLING formation procedures
Developing a mutagenized population: TILLING of any species can be successful based on the frequency of induced mutation in the population to be screened. A wide variety of physical, chemical and biological mutagenic methods can be used to generate genetic alterations.
The most commonly mutagenic agents are chemical class such as:
• Alkylating agents such as Ethyl Methyl Sulfonate (EMS) Nethyl- N-nitrosourea (ENU).
• Diaminating agents including nitrous and nitroso.
• Guanidine haydloxylating agents including hydroxylamine and other chemicals with unknown mechanism of action such as Di Epoxy Butane (DEB) and sodium azide.
EMS is an extremely efficient mutagen that is commonly used in plants to create mutant population. EMS produces random mutation in genetic materials by nucleotide substitutions specifically by guanine alkylation this typically produces only point motions in the DNA sequence or genome. This mutagen induces a large number of recessive species in plants [17,18].
According to first discovery or developed TILLING to screen EMS induced point mutation in Arabidobsis thaliana. They soaked seeds for 20 milli molar-40 milli molar EMS for 10 hours-20 hours and grew them to obtain F1 plants [2,3]. Since F1 plants are chimeric for mutations they self-fertilized F1's to produce F2 line for DNA isolation.
The general protocol for the creation of a TILLING platform in plants includes the following steps:
Creation of mutated populations
• Chemical mutagenesis.
• Development of M1 and M2 generations.
• DNA extraction from individual M2 plants.
• Creation of DNA pools of 5-8 M2 plants.
• Setting up an M3 seed bank.
Detection of mutations in a targeted sequence
• Polymerase Chain Reaction (PCR) amplification of the targeted DNA segment using pooled DNA as a template.
• Detection of mutations using different procedures, e. g. cleavage by specific endonuclease, Denaturing High- Performance Liquid Chromatography (DHPLC) or highthroughput sequencing.
• Identification of the individual M2 plant carrying the mutation.
• Sequencing the target gene segment to confirm the mutation and to determine the type of nucleotide change.
Analysis of the mutant phenotype
A mutated population becomes a TILLING platform when the DNA samples and seeds collected from a large M2 population are archived and put into databases. Usually, platforms of 3,000-5,000 M2 individuals are created, although larger populations that include 10,000 plants have also been reported. Almost all TILLING populations were developed using chemical mutagens, among them, the alkylating agent (EMS) was most often applied. The great mutagenic potential of chemical agents has been proven by the high density of mutations reported for established TILLING populations (Figure 1).
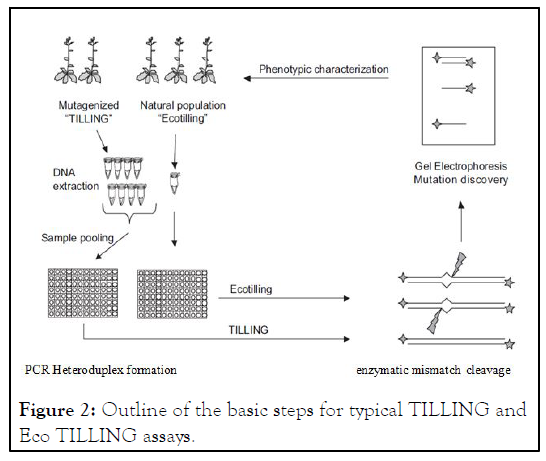
Figure 1: Schematic depicting the TILLING strategy applied to a plant such as arabidopis.
The EMS induced point mutation can be identified by TILLING technique. This is a throughput approach which can now be used to detect single base substitutions in a target or genotype Single Nucleotide Polymorphisms (SNPs). The challenge is in chemical mutagenesis is to determine an ideal does of mutagen which requires a considerable amount of trial and error [19,20].
The main drawback of chemical mutagenesis is difficulty of identifying the mutant phenotype caused by a single base change in a large genome. In addition the resulting damage caused by mutagens can be severe. If the mutation causes deletion, insertion and re arrangement, the mutated allele can lose its functions. The effect of mutagenesis can be homozygous or heterozygous. Homozygous mutations a gene can lead to lethality or sterility. The most desirable mutations are heterozygous mutations.
The chemical mutagen EMS can induce single base change in coding regions resulting in silence, nonsense or miss-silence mutations. The silent mutation does not alter any encoding amino acid so the effect will be silent, whereas missense or nonsense mutations are deleterious. While missense mutation will change the amino acid in the protein by a particular codon change, a nonsense mutation may result in a premature codon stop or a non sense codon in in the transcribed mRNA, and possibly a truncated, and often nonfunctional protein product. Since creating mutant population is particularly difficult for vegetative propagated crops and plants requiring long time for reproduction, eco TILLING is the method of choice to identify natural variations in these plants.
DNA is collected from a mutagenized population (TILLING), or a natural population (Eco TILLING). For TILLING, DNAs from up to eight individuals are pooled. Typical Eco TILLING assays do not use sample pooling, but pooling has been used to discover rare natural single-nucleotide changes. After extraction and pooling, samples are typically arrayed into a 96 well format. The target region is amplified by PCR with gene specific primers that are end labeled with fluorescent dyes. Following PCR, samples are denatured and annealed to form heteroduplexes that become the substrate for enzymatic mismatch cleavage. Cleaved bands representing mutations or polymorphisms are visualized using denaturing polyacrylamide gel electrophoresis. Plants with mutations predicted to affect protein function can be carefully analyzed for phenotypic abnormalities (Figure 2).
Figure 2: Outline of the basic steps for typical TILLING and Eco TILLING assays.
DNA preparation and pooling
Sample pooling will directly affect the efficiency and cost of mutation discovery. With similar false positive and false negative discovery rates, screening four samples pooled together will take approximately twice as long and cost twice as much as screening a pool of eight samples. Factors that affect the ability to pool include the quality of genomic DNA, the accuracy of sample quantification, and the method used for SNP discovery. At the STP, all samples are currently pooled eight-fold. The STP has most often used two basic pooling strategies. For large-scale services, we typically use a one dimensional pooling strategy where each individual sample is represented in only one pool. When a mutation is identified in a pool of eight individuals, each member of the pool is then screened independently to identify the individual harboring the mutation.
The other approach is to pool samples two dimensionally such that each sample is present in two unique pools. Although duplicating each sample reduces the throughput of detection in pools by half, the sample harboring the mutation is unambiguously determined in the pool screening step, so that there is no need to screen individual samples as is done in the one-dimensional strategy. In addition, because two dimensional pooling involves screening each sample with two-fold coverage, potential false positive and false negative errors are minimized at the initial screening step, rather than when individuals are screened in the second step with one dimensional pooling. The current approach for STP is to perform small-scale pilots using two dimensional pooling, where error rates are unknown. Before moving to a large scale operation such as a public TILLING service, the advantage of higher throughput using one dimensional pooling is weighed against the advantage of one step determination and a decision is made on a case-by-case basis.
Mutation discovery: SNP discovery technologies include arraybased methods, denaturing HPLC, mass spectroscopy, denaturing gradient capillary electrophoresis and enzymatic mismatch cleavage. In theory, any accurate SNP discovery method can be used for TILLING. In practice, the method must be both robust and cost effective. Given that the highest density of induced point mutations yet reported for TILLING a diploid species is ~1 mutation/250 kb, screening several thousand mutant individuals will likely be required to ensure a high probability of identifying at least one deleterious mutation. Because of this limitation, whole genome scanning methods such as SNP chips and polony based sequencing are too error-prone to be cost-competitive now.
Application of TILLING and Eco TIILING in plants
Since the inception of TILLING, this method has been widely used for the study of functional genomics in plants, especially for the model plant Arabidopsis thaliana. Reported that the Arabidopsis TILLING Project (ATP), which was set up and introduced as a public service for the Arabidopsis community, had detected 1,890 mutations in 192 target gene fragments. Heterozygote mutations were detected at twice the rate of homozygote mutations. Therefore, the mutational density for treatment of Arabidopsis with EMS was approximately 1 mutation/300 kb of DNA screened with these mutations distributed throughout the genome. The numerous mutations in Arabidopsis thaliana that have been identified via TILLING have provided an allelic series of phenotypes and genotypes to elucidate gene and protein function throughout the genome for Arabidopsis researchers.
Although TILLING was originally designed for and applied mainly in Arabidopsis in the early years of its inception, it has been demonstrated to be an extremely versatile approach compared to many other reverse genetic approaches. This method has proven to be successful to rapidly identify variant genotypes and determine gene function in plants that are diploid and have relatively small genomes such as Arabidopsis. Additionally, it also can be easily applied to other crop plants with very large genomes that are further complicated by various ploidy levels such as wheat.
EcoTIILING is a molecular technique that is similar to TILLING, except that its objective is to uncover natural genetic variation as opposed to induced mutations. Many species is not amenable to chemical mutagenesis; therefore, EcoTIILING can aid in the discovery of natural variants and their putative gene function. This approach allows one to rapidly screen through many samples with a gene of interest to identify naturally occurring SNPs. The method has proven to be successful to detect DNA polymorphisms including variations in satellite repeat number. Furthermore, in highly heterozygous outcrossing species, EcoTIILING can be used to determine heterozygousity levels within a gene fragment. Another valuable application of EcoTIILING is mining for variation in resistance genes to help speed up the process of identifying alleles that could provide immunity to various diseases. Allelic variation had identified by using EcoTIILING in mlo and Mla resistance genes of Hordeum vulgare (barley). These genes are involved in defending the plant from the fungal pathogen that causes powdery mildew. EcoTIILING can be a good technique to employ especially when working with a well established population with thoroughly characterized morphological data.
Crop breeding: Conventional mutation breeding, either by radiation or by chemical treatment, has a proven influence on production of many varieties, including high yielding rice, barley and wheat, etc. Unlike conventional mutation breeding in which the mutation frequency is unknown or estimated from mutations conveying a visible phenotype, TILLING provides a direct measure of induced mutations. Besides, it allows not the prompt, parallel selection of numerous genes but also a forecasting of the number of alleles that will be recognized based on the mutation frequency and library size.
Advantages and Disadvantages of TILLING and Eco TIILING
TILLING is a non-transgenic, high throughput reverse genetic approach. This technique unlike other SNP detection methods provides the approximate location within a few base pairs of the induced mutation, which allows targeted sequencing in the area of the induced mutation opposed to sequencing the entire fragment. Since chemical mutagenesis produces a range of various mutations throughout the genome such as nonsense, splice site, and missense, all of which potentially can affect the protein structure and the resulting phenotype, it has been used for decades to obtain mutants for genetic studies. Therefore, through mutagenesis one can obtain partial loss or complete loss of function and new novel functions, which can provide valuable insight into the true role of a gene in a species of interest. One of the main advantages of TILLING is the amount of time and money this method can potentially save by not requiring sequencing of all individuals in a population to mine for frequent or rare SNPs. TILLING has been demonstrated to be sensitive enough to detect induced mutations and naturally occurring SNPs, as well as the detection of heterozygotes.
Eco TIILING reduces the time and effort for SNP discovery generally required by weeding out identical haplotypes. Therefore, this method does not require one to sequence all individuals in a population to identify polymorphisms, which can be a burdensome expense and time consuming [21]. It also has the advantage of detecting multiple polymorphisms in a single fragment because CELI will digest only a small proportion of the heteroduplexes at a single position. This technique has not been as widely employed as TILLING.
One of the potential disadvantages of Eco TIILING is that when the number of polymorphic sites is high for a gene or PCR fragment of interest across all samples in the population, then an eight fold pooling strategy requires much more labor and time to identify SNPs. This is because eight fold pooling (with a high SNP frequency) would require remixing of numerous pools to locate positive individuals. The other disadvantage is the initial large expenses of TILLING and Eco TILLING experiments are the use of robotic equipment and the purchase of automated sequencers such as the LI-COR DNA analyzer commonly used for cleaved fragment detection. However, these products are not essential and experiments can be carried out without the use of robotics.
Nucleotide sequence variation is mainly determine heritability of phenotypic differences and has been exploited by human for the improvement of crops since the starting of domestication. Variation either can be natural, from divergence populations, or induced through treatment application with mutagens. For decades, mutations are created by treatment with the application of the same chemical mutagens have been employed for TILLING successfully.
TILLING (Targeting Induced Local Lesions in Genomes) is a non transgenic reverse genetic technique that is suitable for most plants whereas, EcoTIILING, an expansion of TILLING technique, can be used to discover point mutations or polymorphisms in natural populations. TILLING formation consists of three main steps namely; Development of a mutagenize, DNA preparation and, mutation discovery.
TILLING and Eco TILLING are important for determining the range of variation for genetic mapping based on linkage analysis. In these techniques, if a mutation is detected in a pool, the individual DNA samples that went into the pool can be individually analyzed to identify the individual that carries the mutation. Once this individual has been identified, its phenotype can be determined. These techniques work with good results, even if a population contains preexisting mutations that would compromise SNP discovery by other methodologies.
This review describes how mutagens, particularly chemical mutagenesis is becoming a powerful tool, especially for reverse genetics in plant species by using the TILLING approach. These new screening methods can be applied to several plant species, whether small or large, diploid or allohexaploid in nature. They may serve as rapid approaches to identify the induced and naturally occurring variation in many species. Eco TIILING relies on naturally occurring mutations rather than induced mutations, using advanced technologies similarly with TILLING to find and use desirable natural gene variants.
Citation: Mekonen AA, Achenef G (2022) Application of TILLING and Eco TILLING in Crop Improvement. J Agri Sci Food Res. 13:504.
Received: 21-Jul-2022, Manuscript No. JBFBP-22-18517; Editor assigned: 23-Jul-2022, Pre QC No. JBFBP-22-18517; Reviewed: 06-Aug-2022, QC No. JBFBP-22-18517; Revised: 31-Oct-2022, Manuscript No. JBFBP-22-18517; Published: 17-Mar-2023 , DOI: 10.35248/2593-9173.23.14.138
Copyright: © 2022 Mekonen AA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.