Journal of Probiotics & Health
Open Access
ISSN: 2329-8901
ISSN: 2329-8901
Research - (2019)Volume 7, Issue 4
The human intestinal microbiota may be considered as a post-natally acquired organ composed of a large diversity of bacteria with different functions on human health. Probiotics are widely used to improve gut functionality and immune system responses. The aim of this study was to evaluate the capability of the bacterial strains Bifidobacterium breve BR03 (DSM 16604) and Lactobacillus plantarum LP01 (LMG P-21021) to induce an in vitro immune response in the peripheral blood mononuclear cells (PBMCs) of healthy adult volunteers and to modify the state of oxidative stress and intestinal permeability of in vitro cell models. Specifically, the analysis was conducted on PBMCs after different stimulation times in order to analyze both cells involved in innate immunity and those responsible for acquired immunity and to evaluate the oxidative stress, and on Caco-2 cell line as an intestinal epithelium model.
Immune system modulation; Gut probiotics; Mucosal adhesion; Intestinal permeability; Oxidative stress
The immune system of vertebrates and, therefore, of humans is an extraordinary integrated network of chemical and cellular agents which developed in the course of evolution to defend the body from any form of chemical trauma or infective attack, thus maintaining the homeostasis of the body itself [1].
Two types of immunity exist based upon the method of recognition of the antigens: Aspecific or innate immunity and specific or acquired immunity [2,3].
Innate immunity is a rapidly evolving field with novel cell types and molecular pathways being discovered and paradigms changing continuously. This branch of immunity involves both cell-dependent mechanisms (e.g. phagocytosis and cytotoxicity) mediated by cellular subtypes, namely phagocytic cells, dendritic cells and natural killer cells, responsible for the first line defense against aggressions [4], and numerous secreted factors, including complement factors [5,6], interferons, alarmins [7,8], cytokines/ chemokines [9], chitinases/chitinase-like proteins [10], acutephase proteins, proteases, and other less-categorized molecules. Innate immune defenses in mammals encompass virtually all tissues, particularly barrier surfaces such as the skin or the mucosal surfaces of the respiratory and gastrointestinal tract [11]. The innate immune system can “sense” tissue damage, infection, or genotoxic stress through germline-encoded receptors (e.g. pattern recognition receptors [PRRs] such as toll-like receptors [TLRs]) [12].
On the other side, acquired immunity includes soluble factors (cytokines) and cells (T lymphocytes, B lymphocytes and accessory cells) responsible for a more powerful and focused but slower defensive response (virtually capable of recognizing any form of antigen) [13]. Unlike innate immunity, the acquired response was selected by evolution for its capacity to dynamically adapt to the variability of environmental agents recognized as dangerous to the body [14].
Cytokines are a heterogeneous group of soluble proteins of low molecular weight (very often glycoproteins) which mediate the interactions between cells of the immune system and other cells in the body [15]. Cytokine signaling is indispensable for regulatory T-cell (Treg) development in the thymus, and also influences the homeostasis, phenotypic diversity, and function of Tregs in the periphery [16].
Anyway, an unbalanced immune response may lead to severe inflammation and uncontrolled tissue damage and disease [17].
The mucosal membrane of the intestine, with an overall surface area of approximately 300 m2, is constantly challenged by the enormous number of antigens from food, from the intestinal microbiota, and from inhaled particles that also reach the gut at least partially [18]. It is not surprising therefore that approximately 80% of the immune system is located in the intestinal tract area and is particularly prevalent in the small intestine [19].
Sensing of the intestinal microbiota by the host mucosal immune system plays significant roles in maintaining intestinal homeostasis and inducing systemic protective responses. Thus, manipulation of the intestinal microbiota is a potential alternative approach for maintaining health and preventing or treating diseases [20].
Probiotics are widely used to rebalance microbiota composition, thus improving gut functionality and immune system responses [21].
One of the major mechanisms of probiotic action is through the regulation of the host immune response and intestinal mucosal defences [22]. These include blocking pathogenic bacterial effects by synthesizing bactericidal substances and competing with pathogens and toxins for adherence to the intestinal epithelium [23].
Recent research on the molecular biology and genomics of Lactobacillus and Bifidobacterium has focused on the interaction with the immune system and potential as a biotherapeutic agent in cases of antibiotic-associated diarrhoea, travellers’ diarrhoea, pediatric diarrhoea, inflammatory bowel disease (IBD) and irritable bowel syndrome (IBS) [24-26].
There is good evidence that certain strains of Lactobacilli and Bifidobacteria can influence immune regulation, particularly through a modulation of the balance between pro-inflammatory and anti-inflammatory cytokines [27].
For more than two decades, immunologists have been using the so-called Th1/Th2 paradigm to explain most of the phenomena related to adaptive immunity [28].
The Th1/Th2 paradigm implied the existence of two different, mutually regulated, CD4+ T helper subsets which play a central role in modulating immune responses: Th1 cells producing IFN- γ drive cell-mediated immune responses involved in tissue damage and fight infections caused by intracellular parasites; and Th2 cells secreting interleukin (IL)-4, IL-5, IL-13 and IL-25 mediate IgE production, eosinophilic inflammation, allergy and the protection against helminthic parasite infections. A third member of the T helper set, IL-17-producing CD4+ T cells, now called Th17 cells, was recently described as a distinct lineage that does not share developmental pathways with either Th1 or Th2 cells. The Th17 subset has been linked to autoimmune disorders, being able to produce IL-17, IL-17F and IL-21 among other inflammatory cytokines. Th17 cells are also critical to enhance host protection against extracellular bacteria and fungi, which are not efficiently cleared by Th1 and Th2 responses [29,30]. IL-17 has a well-recognized role in immune surveillance at mucosal and barrier surfaces, but also has been progressively more implicated as a driver of immunopathology in situations of autoimmunity and chronic inflammation [31].
Interestingly, it has been reported that there is not only a crossregulation among Th1, Th2 and Th17 effector cells but there is also a dichotomy in the generation of Th17 and T regulatory cells. Therefore, Treg and Th17 effector cells arise in a mutually exclusive fashion, depending on whether they are activated in the presence of TGF-β or TGF-β plus inflammatory cytokines such as IL-6. Therefore, the Th1/Th2 paradigm has now developed into the new Th1/Th2/Th17 paradigm [32,33].
Another important aspect to be taken into account is the complex interplay between immune responses and potential alteration of gut permeability. Inflammatory cytokines regulate tight junctions (TJ) protein expression and function, as well as overall mucosal barrier properties. The most studied cytokine known to cause barrier dysfunction is TNF-α. Anti-TNF-α therapy restores gut barrier function in Crohn’s disease (CD) [34]. However, TNF-α also helps regulate intestinal epithelial cell death and injury or repair [35].
A defective mucosal barrier may result in increased intestinal permeability which promotes the exposition to luminal content and triggers an immunological response that promotes intestinal inflammation. In clinical practice, several studies have documented that changes in intestinal permeability can predict Irritable Bowel Diseases (IBD) course. Functional tests, such as the sugar absorption tests or the novel imaging technique using confocal laser endomicroscopy, allow an in vivo assessment of gut barrier integrity [36].
The rationale for the use of probiotics in IBD is the dysbiosis that characterizes these diseases. The mechanisms of their effect in Ulcerative Colitis (UC) have not been fully understood but probably, along with direct anti-inflammatory effects, they may strengthen mucosal barrier [37,38] and reduce intestinal permeability once again upregulating TJs proteins [39].
The aim of this study was to evaluate the capability of the probiotic strains Lactobacillus plantarum LP01 (LMG P-21021) and Bifidobacterium breve BR03 (DSM 16604) to induce an in vitro immune response in the peripheral blood mononuclear cells (PBMCs) collected from healthy adult volunteers and to modify the state of oxidative stress and intestinal permeability of appropriate in vitro cell models.
Bacterial cultures and growth conditions
The strains Lactobacillus plantarum LP01 (LMG P-21021) and Bifidobacterium breve BR03 (DSM 16604) were grown overnight at 37°C in de Man, Rogosa and Sharpe (MRS) broth. Subsequently, 1 ml of culture was transferred to 10 ml of fresh MRS and incubated at 37°C for 6 h until mid-log phase. It is very important, in fact, that the bacterial cells are harvested during the exponential phase for the following evaluations. The growth curve is derived by the measure of the optical density (OD) at 600 nm of a liquid broth culture after specific intervals of time from the inoculum. Bacterial cells were washed twice with sterile phosphate buffered saline (PBS, pH 7.2).
The physiological state and the number of cells was determined by cytofluorimetric analysis, utilizing the commercial kit “Cell Viability Kit with liquid beads” (Becton Dickinson) according to the instructions provided by the manufacturer to accurately quantify the number of live, dead, and total bacteria in a sample. This analysis is based on the two fluorescent dyes thiazole orange (TO) and propidium iodide (PI) (Life Technologies). The first is able to bind with highest affinity to double-stranded (ds) DNA, while the latter binds to DNA by intercalating between the bases with little or no sequence preference and with a stoichiometry of one dye per 4-5 base pairs of DNA. When excited by 488 nm of laser light, it can be detected with 562-588 nm band pass filter.
Live cells have intact membranes and are impermeable to dyes such as PI, which leaks into cells with compromised membranes. TO is a permeant dye and enters all cells, live and dead, to varying degrees. The fluorescent signal from TO in viable cells allows their enumeration even when debris in the cell preparation contaminates a scatter gate around the cells. In this way, the combination of these two dyes provides a rapid and reliable method for discriminating live and dead eukaryotic and prokaryotic cells, including peripheral blood mononuclear cells (PBMCs).
The cells were then brought to the optimal concentration established in preliminary experiments and employed in the stimulation tests described below.
Preparation of PBMCs
The PBMCs are the mononuclear cellular component of blood, consisting of T lymphocytes (about 75%), B lymphocytes, monocytes and natural killer (NK) cells.
PBMCs were separated by Ficoll-Hypaque (Lymphoprep, ρ=1.077 g/ml) density gradient centrifugation from freshly collected, leukocyte-rich buffy coats supplemented with heparin as anticoagulant and provided by the local Hospital Transfusion Service. Subjects were eligible if they were in good general health and were not currently taking medications, probiotics, and other supplements.
The concentration of separated cells was determined by cellular count in Bürker chambers. The cells were washed twice with RPMI-1640 (Invitrogen) and then diluted to a concentration of 2 × 106 cells/ml of RPMI-1640 culture medium supplemented with 10% heat-inactivated fetal calf serum (FCS, Gibco), 1% Lglutamine and 5mM Hepes (Sigma-Aldrich).
PBMCs were incubated at a concentration of 1 × 105 /100μl/ well in 96 wells U-bottomed microplates for the co-culture experiment in the presence or absence of different stimulants at 37°C under 5% CO2 for a time up to 5 days.
Cell viability (cytotoxicity test)
To assess potential bacterial toxicity towards PBMCs, cell viability was evaluated using the methylthiazolyldiphenyltetrazolium bromide (MTT) assay. The immune cells (1 × 105 units) were challenged for 24 h with bacterial strains (1 ×105 cells) under investigation; then, the medium with the probiotic strains was replaced by the MTT assay solution (1 mg/ml; 2 h, 37°C in the dark). The supernatant was removed and DMSO was added in order to dissolve the purple formazan; the absorbance was then read at 580 and 675 nm. The positive control (CTR+) used was based on Caco-2 cells stimulated with DMSO.
PBMCs stimulation and cytokines quantification: After separation, the PBMCs were stimulated with the bacterial strains for 1 and 5 days.
Internal controls for each experiment were represented by:
Negative control: PBMCs alone
1 day control: PBMCs stimulated with 1 μg/ml of Lipopolysaccharide (LPS; Escherichia coli, serotype 055:B5, Sigma-Aldrich).
5 day control: PBMCs stimulated with 1 μg/ml of Phytohemagglutinin (PHA-P; Sigma-Aldrich).
At different times of analysis, the cultures were centrifuged at 1,500 rpm for 10 minutes. The supernatants were stored at -20°C until analysis, while the cells were utilized for the following experiments.
Cytokines release in the supernatants was determined by sandwich enzyme-linked immunosorbent assay (E.L.I.S.A.) using commercial kits (Bender MedSystems) according to the manufacturer’s instructions.
Superoxide anion (O2-) production
PBMCs (1 × 106 cells/plate) were treated for 24 h with the two probiotic strains. O2- production was evaluated by the superoxide dismutase-sensitive cytochrome C reduction assay and expressed as nmol reduced cytochrome C/106 cells/30 min, using an extinction coefficient of 21.1 mM. To avoid interference with spectrophotometrical recordings, cells were incubated with RPMI-1640 devoid of phenol red and fetal bovine serum (FBS). The positive control (CTR+) employed was represented by Caco-2 cells stimulated with LPS.
Adhesion assay
For the adhesion assay, Caco-2 monolayer was washed twice with PBS pH 7.4 before use. The suspension of BR03 or LP01 (106 CFU/ml) in Dulbecco ’ s Modified Eagle Medium (DMEM, Gibco) was added to the apical wells of Caco-2 monolayer in a total volume of 0.5 ml [40]. The inoculated tissue culture plate was incubated under 5% CO2 at 37°C for 1 h. Unbound bacterial cells were analyzed and counted by flow cytometry with a BD ™ Cell Viability Kit (according to the manufacturer ’ s instructions) based on analytical method ISO 19344:2015 (E)- IDF 232:2015 (E).
Trans-epithelial electrical resistance (TEER) assay
TEER was employed to measure the integrity of tight junctions among the intestinal epithelial cells according to a method described previously [41]. Caco-2 cells (Anemocyte) were seeded into the cell culture inserts (0.4 μm) in 2.5 ml aliquots per well with a concentration of 5 × 105 cells/ml. DMEM supplemented with 10% FBS, 1% mixture penicillin-streptomycin solution and 1% non-essential amino acid solution was added to the basal compartment of each well. The cells were grown at 37°C in an atmosphere of 5% CO2 and 95% air. The culture medium was refreshed every 2 days. After 21 days of culture, the integrity of the monolayer was evaluated by measuring the TEER with an Epithelial Voltohmmeter (EVOM) (World Precision Instruments). The EVOM system has a measurement range of 1-9,999 Ω with a 1 Ω resolution and uses a pair of electrodes popularly known as a STx2/“chopstick” electrode pair.
TEER at time zero was determined before bacterial samples were added to the monolayer. The culture medium from both the apical and basal compartments was then removed and 0.5 ml of bacterial samples were immediately added to the apical side (108-109 CFU/ml), while 1 ml of medium was put in the basal compartments. The cells were maintained for 24 h at 37°C in an atmosphere composed of 5% CO2 and 95% air. Then, the culture medium and the bacterial samples were removed and the apical portion was washed with 2 ml of Dulbecco’s phosphatebuffered saline (DPBS). Thereafter, in both the apical and basal compartments, 2 ml of culture medium were added and TEER readings were recorded after 1h in order to estimate the potential increase of this parameter. One insert/well was left without bacteria as a negative control. The percentage changes in TEER were expressed according to the following equation:
% change in TEER=((T1-T0)/T0) × 100
Where T0: TEER at time zero, and T1: TEER at the end of the above described protocol. Each experiment was conducted in triplicate.
The TEER has been evaluated using two different stimulation methods.
In the prevention assay, a 1-hour incubation with the two probiotics (3 × 106 cells/well) was performed before the 1-hour pro-inflammatory insult carried by TNF-α and IL-1β at the final concentration of 10 ng/ml at 37°C. At the end of the double stimulation, an evaluation of the bacterial adhesion to the epithelial layer has been performed.
In the restoration assay, the probiotic stimulus was applied after the inflammatory challenge in order to quantify the overall possible protective effects resulting from the two beneficial bacteria.
In both protocols, after the recovery of supernatant samples, the wells have been washed with DPBS and fresh DMEM supplemented with 10% FBS was added. The TEER has been quantified after 24h from the medium change.
The positive control (CTR+) was represented by Caco-2 cells stimulated with TNF-α and IL-1β [42].
Immunomodulation results are expressed as the means ± standard error of the mean (S.E.M.) of at least 3 different independent experiments. Statistical analysis was performed on the basis of the program GraphPad Prism 7 using the paired Student’s t-test. Values of p<0.05 were considered significant.
Stimulation of PBMCs with the bacteria and cytokines quantification
With the aim to understand the underlying mechanisms of the probiotic bacteria effects, we investigated the impact of Lactobacillus plantarum LP01 and Bifidobacterium breve BR03, either alone or associated in a 1:1 ratio, on Th1-, Th2- and Th17-cytokine secretion from PBMCs of healthy subjects.
The results after 5 days of co-incubation are shown in Figures 1-3. B. breve BR03 was a strong inducer of IL-10 synthesis (6-fold increase compared to basal level, p<0.001), while L. plantarum LP01 was able to induce an increase of 3.73 times compared to the control (p<0.001), with the 1:1 ratio positioning more or less in the middle (5.27 increase).
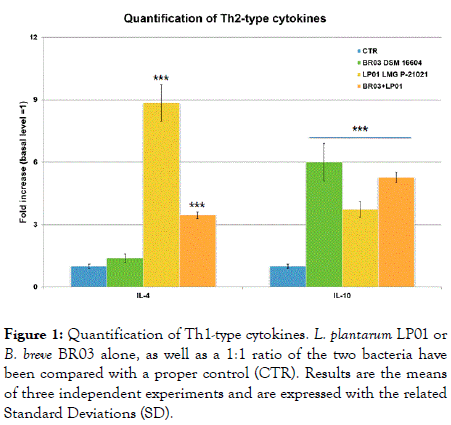
Figure 1. Quantification of Th1-type cytokines. L. plantarum LP01 or B. breve BR03 alone, as well as a 1:1 ratio of the two bacteria have been compared with a proper control (CTR). Results are the means of three independent experiments and are expressed with the related Standard Deviations (SD).
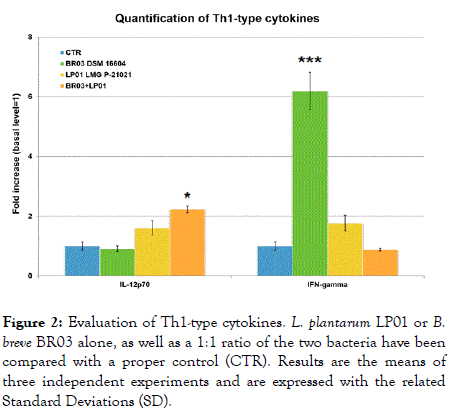
Figure 2. Evaluation of Th1-type cytokines. L. plantarum LP01 or B. breve BR03 alone, as well as a 1:1 ratio of the two bacteria have been compared with a proper control (CTR). Results are the means of three independent experiments and are expressed with the related Standard Deviations (SD).
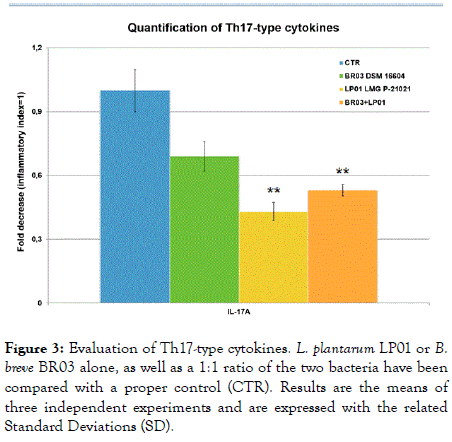
Figure 3. Evaluation of Th17-type cytokines. L. plantarum LP01 or B. breve BR03 alone, as well as a 1:1 ratio of the two bacteria have been compared with a proper control (CTR). Results are the means of three independent experiments and are expressed with the related Standard Deviations (SD).
For what concerns the other Th2-type cytokine, IL-4, the situation is the opposite, meaning that the LP01 strain was a really strong inducer of the cytokine (8.85 times increase compared to CTR, p<0.001), while BR03 alone induced almost no effect on this parameter. Again, the mix positioned itself more or less in the middle (3.45 increase, p<0.001) (Figure 2).
IFN-γ was deeply induced by B. breve BR03 (more than 6 times increase, p<0.001), while LP01 brought an 80% increase (ns). Curiously, the mix showed no effects compared to the control, therefore suggesting that the association of the immunological stimuli brought by the two bacteria may have a reciprocal dampening result.
IL-12p70 recorded a mean scarce induction by the two bacteria used alone, whereas the mix recorded a statistically significant increase (2.23 times, p<0.05) (Figure 1).
It is also interesting to point out that the effects on Th-17 cytokines was based on a general inhibition by the bacteria, with L. plantarum LP01 showing the most relevant impact (57% decrease, p<0.01) (Figure 3).
The secretion of the chemokine IL-8 was significantly reduced by the two probiotics (Figure 4), as well as that of the interferon- α (Figure 5) (p<0.05 for all the samples). The most significant result has been recorded with LP01 strain showing a 39% decrease of interferon-α.
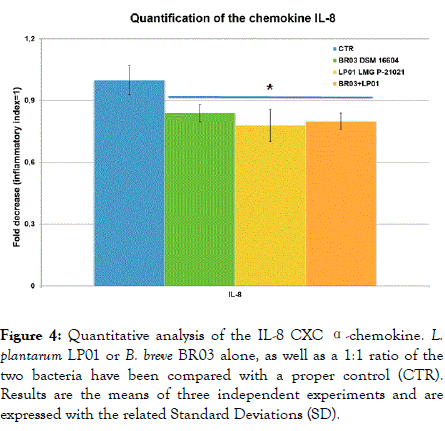
Figure 4. Quantitative analysis of the IL-8 CXC α-chemokine. L. plantarum LP01 or B. breve BR03 alone, as well as a 1:1 ratio of the two bacteria have been compared with a proper control (CTR). Results are the means of three independent experiments and are expressed with the related Standard Deviations (SD).
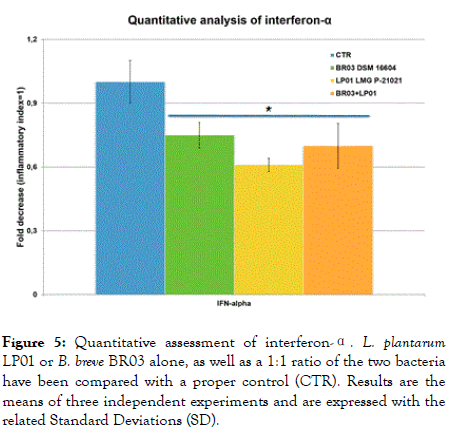
Figure 5. Quantitative assessment of interferon-α. L. plantarum LP01 or B. breve BR03 alone, as well as a 1:1 ratio of the two bacteria have been compared with a proper control (CTR). Results are the means of three independent experiments and are expressed with the related Standard Deviations (SD).
Finally, IL-6 and TNF-α, two important proinflammatory cytokines, were significantly induced especially by L. plantarum LP01 (15.95 times and 47.76 times increase compared with the CTR, respectively; p<0.001). B. breve was a mild inducer of both parameters (3.22 and 5.52 times increase, p<0.001), with the 1:1 ratio again in an intermediate position (Figure 6).
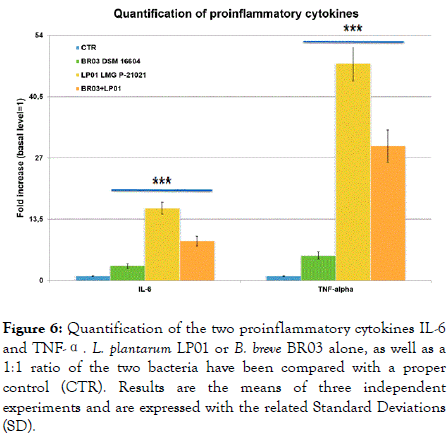
Figure 6. Quantification of the two proinflammatory cytokines IL-6 and TNF-α. L. plantarum LP01 or B. breve BR03 alone, as well as a 1:1 ratio of the two bacteria have been compared with a proper control (CTR). Results are the means of three independent experiments and are expressed with the related Standard Deviations (SD).
The stimulation of production of Th2 cytokines seemed to be inversely related to the production of Th1 cytokines, at least to a certain extent and for some specific cytokines.
In all experiments, stimulation with the mitogen control (PHA) always resulted in values greater than the threshold, confirming that the PBMCs utilized were vital and capable of proliferation.
Cytotoxicity test
The CTR+ sample (Caco-2 cells stimulated with dimethyl sulfoxide-DMSO-in the MTT test) showed 100% mortality, as expected. The probiotic bacteria showed very slight variations compared with the negative control, with only L. plantarum LP01 alone suggesting a very minimal mortality (6 ± 7.5%), even if not to a statistically significant extent (Table 1).
| Test | CTR- | CTR+ | BR03 | LP01 | BR03+ LP01 |
|---|---|---|---|---|---|
| 1. Cytotoxicity test | 100% (vitality) | 0% (death) | 104 ± 8.3% | 94 ± 7.5% | 100 ± 8% |
| 2b. Prevention test | 100% (total membrane integrity) | 40 ± 4.4% (damage) | 95 ± 7.6% | 101 ± 8.2% | 93 ± 7.4% |
| 2c. Restoration test | 100% (total membrane integrity) | 40 ± 4.4% (damage) | 94 ± 7.5% | 100 ± 9% | 91 ± 7% |
| 3. Adhesion test | 95 ± 8% | 3 ± 0.25% | 37 ± 3% | ||
| 4. Superoxide anion (O2-) production | 0 (basal level) | 1 (inflammatory level) | 0.39 ± 0.041 | 0.35 ± 0.028 | 0.35 ± 0.045 |
Table 1: Parameters of phenotypic characterization of the strains. CTR-: unstimulated cells used as negative control; CTR+: DMSO-stimulated cells in the MTT test (1), TNF-α+ IL-1β-stimulated cell in the TEER assay (2a, 2b) and LPS-stimulated cells in the ROS production assay (4) as positive controls. No controls were employed in the adhesion test (3). Both controls (CTR+ and CTR-) were compared with the two probiotic bacteria, used either alone or in a 1:1 ratio.
Superoxide anion (O2-) production
Defining the basal level as 0 and the inflammatory level as 1, the bacteria positioned closer to the former. B. breve BR03 recorded a score of 0.39 ± 0.041, while L. plantarum LP01 had an impact equal to 0.35 ± 0.028. The mix gave a result very close to LP01 strain (Table 1).
Adhesion assay
The two bacteria showed opposite results, with L. plantarum LP01 behaving as a very poor adhesive strain (3 ± 0.25%), while almost the totality of BR03 cells were found adhered to Caco-2 cells (95 ± 8%). As it could be expected, the mix provided a 37 ± 3% result (Table 1).
Trans-epithelial electrical resistance (TEER) assay
In both the prevention and the restoration test the two controls gave the expected results, that is total membrane integrity in the negative control and 40 ± 4.4% in the positive control (significant damage). The strain L. plantarum LP01 was able to fully prevent any damage to the tight junctions of the Caco-2 cells in both tests, while B. breve BR03 showed slightly lower results (95 ± 7.6% in the prevention and 94 ± 7.5% in the restoration). Curiously, the mix of the bacteria has proven to be slightly worse than BR03 strain alone (93 ± 7.4% in the prevention and 91 ± 7% in the restoration).
The immunomodulatory activity of a probiotic strain on PBMCs could be roughly divided in three aspects:
• Induction of cell proliferation;
• Modulation of phenotypic expression;
• Production of cytokines and immunoglobulins by B lymphocytes.
Many evidences suggest that probiotic bacteria may have, in a species- or even strain-dependent manner, a potential use as antiinflammatory agents in some chronic inflammatory diseases [43].
On the basis of experimental data, the anti-inflammatory effects of probiotics may be a consequence of their antagonism of potentially pathogenic/proinflammatory endogenous microbiota, the modulation of balance between Th1, Th2 and regulatory T (Treg) cells, the down-regulation of proinflammatory (e.g. IL-12, TNF-α) and/or stimulation of anti-inflammatory (e.g. IL-10) cytokines production. Other effects such as enhanced elimination, modified degradation, permeation and presentation of proinflammatory antigens can play an important role in this sense, especially in the human gut [44].
IFN-γ and IL-12 are well known for the induction of the differentiation of T helper lymphocytes in Th1 cells, which are essential for cell-mediated immunity. IL-4, IL-5 and IL-10 trigger the differentiation of Th2 cells, typically associated with humoral immunity and host defense against extracellular pathogens.
Numerous studies support the view that IL-10 exerts a strong suppressive effect on Th1 lymphocytes, antigen presenting cells and the production of inflammatory mediators [45]. Thus, IL-10 can counteract the production of other cytokines such as IFN-γ or IL-4.
To date pathogenic, probiotic and commensal bacteria are considered to induce different levels of immune response: a strong host response stimulated by pathogens, an intermediate response induced by probiotics, and finally a homeostatic control of the response triggered by commensal bacteria. It should also be remembered that interactions between Pathogen Recognition Receptors (PRR) and ligands are not as specific as those between antigens and antibodies, and ligands for PRR such as Toll-like receptors (TLR) are generally present in repetitive structures to increase avidity. Therefore, some very important and challenging questions concerning immunemediated probiotic activity could be the following [46]:
• Are whole live bacteria essential to promote biological effects on the immune system?
• Can the concept of probiotics be extended to include bacterial-derived molecular bioactive components?
• Moreover, can probiotic molecules be produced also by nonprobiotic bacteria?
Our results highlight the potential usefulness of an association of the probiotic strains L. plantarum LP01 and B. breve BR03 in the treatment and prevention of gastrointestinal inflammatory disorders as well as the reduction of bacterial overgrowth in IBS by promoting Th2- and counteracting Th1-immune response.
A previous human clinical study by Saggioro [47] demonstrated a great efficacy of the same combination of L. plantarum LP01 and B. breve BR03 in relieving pain of the main symptoms typically associated with Irritable Bowel Syndrome (IBS), thus suggesting a strong anti-inflammatory activity mediated by the strains in the human gut.
The simple in vitro model described in our work could be considered as very reliable and predictive of the in vivo interaction of a specific strain with the gut-associated lymphoid tissue (GALT), therefore suggesting the possible usefulness of a microorganism in the treatment of allergies or inflammatory bowel conditions.
Besides the overall effects on the immune system, it is also interesting to point out the ability to help restore the physiological gut barrier mediated by the TJs. This positive effect does not seem to be related to the adhesion of probiotic cells to the inner surface of the intestinal mucosa. As a matter of fact, only B. breve BR03 was able to significantly adhere to the cell line used in the experiment, while both strains, separately and combined demonstrated a strong efficacy in the restoration of a physiological barrier.
Disruption of the gut barrier has been associated with many gastrointestinal diseases, but also with extra-intestinal pathological condition, such as type 1 diabetes mellitus, allergic diseases or autism spectrum disorders [48,49]. Several studies have highlighted the role of probiotics in the modulation and reduction of intestinal permeability, considering the strong influence of gut microbiota in the modulation of the function and structure of gut barrier, but also on the immune response of the host. Probiotics have also a role in the regulation of the function of gut barrier, by altering mucus or chloride secretion but also directly preventing the rearrangement of tight junction proteins after exposure to pathogenic bacteria [50], or restoring a disrupted epithelial barrier [51].
Our results significantly confirm the results previously found in the scientific literature, also extending their attribution to the species Bifidobacterium breve, that could be fruitfully employed in both IBS and chronic gut dysbiosis associated with impaired mucosal permeability like the one reported in coeliac disease [52], thus potentially contrasting the progression of irritable bowel diseases in predisposed patients.
Special thanks are dedicated to our colleague Stefania Nicola, who contributed to the generation of a significant percentage of the scientific results hereby presented, with special reference to the cytokine panel. Nina Vinot contributed with the English proofreading of the manuscript.
The study has been entirely funded by Probiotical S.p.A.
Citation: Amoruso A, Deidda F, Pane M, Mogna L (2019) A Systematic Evaluation of the Immunomodulatory and Functional Properties of Probiotic Bifidobacterium breve BR03 (DSM 16604) Lactobacillus plantarum LP01 (LMG P-21021). J Prob Health. 7:214. DOI: 10.35248/2329-8901.19.7.214
Received: 06-Dec-2019 Accepted: 20-Dec-2019 Published: 27-Dec-2019
Copyright: © 2019 Amoruso A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Sources of funding : The study has been entirely funded by Probiotical S.p.A.