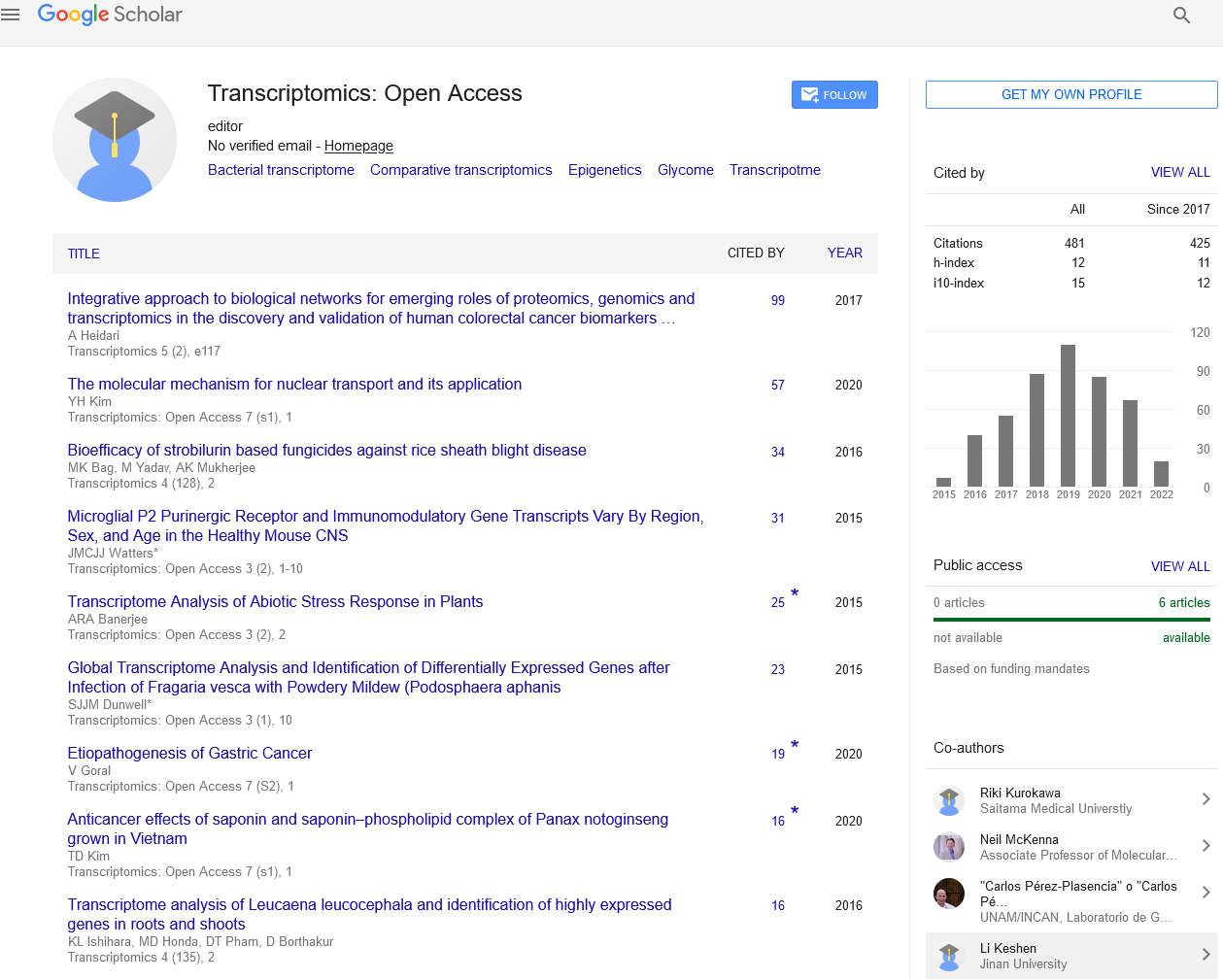
Transcriptomics: Open Access : Citations & Metrics Report
Articles published in Transcriptomics: Open Access have been cited by esteemed scholars and scientists all around the world. Transcriptomics: Open Access has got h-index 13, which means every article in Transcriptomics: Open Access has got 13 average citations.
Following are the list of articles that have cited the articles published in Transcriptomics: Open Access.
| 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
Total published articles |
30 | 32 | 30 | 25 | 24 | 0 | 2 | 4 | 14 | 28 | 6 | 6 |
Research, Review articles and Editorials |
0 | 2 | 1 | 10 | 5 | 0 | 1 | 1 | 9 | 17 | 5 | 6 |
Research communications, Review communications, Editorial communications, Case reports and Commentary |
14 | 40 | 30 | 16 | 8 | 0 | 1 | 2 | 5 | 11 | 1 | 0 |
Conference proceedings |
0 | 0 | 0 | 0 | 0 | 0 | 0 | 50 | 52 | 111 | 0 | 0 |
Citations received as per Google Scholar, other indexing platforms and portals |
42 | 58 | 67 | 78 | 96 | 113 | 96 | 67 | 57 | 6 | 2 | 3 |
| Journal total citations count | 552 |
| Journal impact factor | 2.08 |
| Journal 5 years impact factor | 4.98 |
| Journal cite score | 6.92 |
| Journal h-index | 13 |
| Journal h-index since 2019 | 11 |
Important citations (451)
jambagi s, dunwell jm. identification and expression analysis of fragaria vesca mlo genes involved in interaction with powdery mildew (podosphaera aphanis). journal of advances in plant biology. 2017;1(1):41-55. |
|
wei y, shao x, wei y, xu f, wang h. effect of preharvest application of tea tree oil on strawberry fruit quality parameters and possible disease resistance mechanisms. scientia horticulturae. 2018 nov 18;241:18-28. |
|
zhong y, zhang x, cheng zm. lineage-specific duplications of nbs-lrr genes occurring before the divergence of six fragaria species. bmc genomics. 2018 dec;19(1):128. |
|
miao lx, jiang m, zhang yc, yang xf, zhang hq, zhang zf, wang yz, jiang gh. genomic identification, phylogeny, and expression analysis of mlo genes involved in susceptibility to powdery mildew in fragaria vesca. genet. mol. res. 2016 aug 5;15. |
|
kerry rg, mahapatra gp, patra s, sahoo sl, pradhan c, padhi bk, rout jr. proteomic and genomic responses of plants to nutritional stress. biometals. 2018 apr 1;31(2):161-87. |
|
banerjee a, roychoudhury a. role of beneficial trace elements in salt stress tolerance of plants. inplant nutrients and abiotic stress tolerance 2018 (pp. 377-390). springer, singapore. |
|
banerjee a, wani sh, roychoudhury a. epigenetic control of plant cold responses. frontiers in plant science. 2017 sep 21;8:1643. |
|
banerjee a, roychoudhury a. epigenetic regulation during salinity and drought stress in plants: histone modifications and dna methylation. plant gene. 2017 sep 1;11:199-204. |
|
kumar v, khare t, arya s, shriram v, wani sh. effects of toxic gases, ozone, carbon dioxide, and wastes on plant secondary metabolism. inmedicinal plants and environmental challenges 2017 (pp. 81-96). springer, cham. |
|
malik ah, ashraf n. transcriptome wide identification, phylogenetic analysis, and expression profiling of zinc-finger transcription factors from crocus sativus l. molecular genetics and genomics. 2017 jun 1;292(3):619-33. |
|
das s, kar rk. abscisic acid mediated differential growth responses of root and shoot of vigna radiata (l.) wilczek seedlings under water stress. plant physiology and biochemistry. 2018 feb 1;123:213-21. |
|
devi el, kumar s, singh tb, sharma sk, beemrote a, devi cp, chongtham sk, singh ch, yumlembam ra, haribhushan a, prakash n. adaptation strategies and defence mechanisms of plants during environmental stress. inmedicinal plants and environmental challenges 2017 (pp. 359-413). springer, cham. |
|
wani sh, dutta t, neelapu nr, surekha c. transgenic approaches to enhance salt and drought tolerance in plants. plant gene. 2017 sep 1;11:219-31. |
|
sah sk, reddy kr, li j. abscisic acid and abiotic stress tolerance in crop plants. frontiers in plant science. 2016 may 4;7:571. |
|
vana dr. the molecular crosstalk: oxidative stress, genomic instability and neurodegenerative disorders. biomedical research. 2017;28(14). |
|
borriss r. phytostimulation and biocontrol by the plant-associated bacillus amyloliquefaciens fzb42: an update. inbacilli and agrobiotechnology 2016 (pp. 163-184). springer, cham. |
|
karimi r, vahedi m, pourmazaheri h, balilashaki k. advances in biomedicine and pharmacy. |
|
kothari v, patel p, joshi c. modulation of microbial quorum sensing: nanotechnological approaches. indesign of nanostructures for versatile therapeutic applications 2018 (pp. 523-563). |
|
ramesh sv, williams s, kappagantu m, mitter n, pappu hr. transcriptome-wide identification of host genes targeted by tomato spotted wilt virus-derived small interfering rnas. virus research. 2017 jun 15;238:13-23. |
|
singh i, smita s, mishra dc, kumar s, singh bk, rai a. abiotic stress responsive mirna-target network and related markers (snp, ssr) in brassica juncea. frontiers in plant science. 2017 nov 21;8:1943. |
|