Journal of Clinical & Experimental Dermatology Research
Open Access
ISSN: 2155-9554
ISSN: 2155-9554
Research Article - (2023)Volume 14, Issue 2
Introduction: The microbiome represents one of the different factors involved in the pathogenesis of acne. Negative effects on skin microbiome were described after the use of specific acne treatments (like isotreinoin and benzoyl peroxide) or aggressive cleansers. Considering that skin cleansing protocols represent an important step in acne care, the use of a cleanser that preserve or restore the microbiome balance represents a key strategy in the treatment of acne. In this context, a prospective, proof of concept, open label trial was conducted to evaluate the effect of a cleanser containing Aloe Vera and Biopep15 (an oligopeptide with antibacterial and anti-inflammatory activities) on skin microbiome in subjects with acne-prone skin.
Materials and methods: Fifteen subjects (11 women and 4 men, mean age 23 ± 3 years), with acne-prone skin, were enrolled in the study. The cleanser was applicated on the face twice a day (morning and evening) for 28 consecutive days. The skin acceptability (clinical signs and sensations of discomfort) and the effect on skin microbiome (by means of skin DNA sequencing) were evaluated after 28 days of product used.
Results: The cleanser showed very good skin acceptability: No clinical signs or sensations of discomfort were observed during the treatment. No alteration in microbial richness and in microbial diversity (evaluated by OTUs, Chao and Shannon Indexes) was observed after 28 days of treatment, although a slight not significant increase in microbial diversity was observed. In the present study a decrease in Actinobacteria (-0.38 ± 23.04%) and Firmicutes (-7.14 ± 17.06%), and an increase in Proteobacteria (5.48 ± 17.75%) and Bacteroidetes (2.41 ± 4.72%, p<0.05), although generally not statistically significant, was observed at the end of the treatment. According to the literature, Actinobacteria and Firmicutes are generally increased in acne patients while Proteobacteria are usually under-represented in the acne skin patients. At genus level a statistical decrease of Corynebacterium (-1.86 ± 3.15%, p<0.05) and a decrease of Staphylococcus and Streptococcus (-1.50 ± 15.89% and -2.93 ± 11.10%, respectively), both generally abundant on acne skin, were observed.
Conclusion: The cleanser evaluated in this study was well tolerated with preserving and restoring effects on microbiome balance in acne patients. Therefore, it could be used as adjuvant product for effective acne treatments like benzoyl peroxide and oral isotretinoin which could have negative effects on skin microbiome.
Skin microbiome; Acne vulgaris; cleansing protocol; Skin dysbiosis
Acne is a chronic inflammatory skin condition that involves sebaceus rich sites [1]. The pathogenesis of acne involved an interplay of several factors such as increasing sebum production and inflammatory mediators, and follicular keratinization of the pilosebaceous ducts [2]. In addition, the interaction between microbiome and the skin functions and immunity seems to play an important role in the pathogenesis of this disease [3]. The skin microbiome is composed by commensal and pathogenic bacteria [4]. The commensal bacteria (transient or resident), contribute to create a healthy and balance ecosystem [4]. A microbial imbalance (dysbiosis) has been suggested to be involved in the development of inflammatory acne [5]. The main known bacteria involved in the development of acne is Cutibacterium acnes, that seems to increase lipid production and starts inflammatory response through Toll-Like Receptor 2 (TLR2) activation [6]. However, other skin bacteria, such as Staphylococcus epidermidis, seem to play an important role in the acne development [7]. The most commensal skin bacteria could be classified into the following four phyla: Actinobacteria (51.8%), Bacteroidetes (6.3%), Firmicutes (24.4%), and Proteobacteria (16.5%) [8,9]. The microbial population could also be divided based on the skin area considered: Sebaceous or oily (face, chest and back); moist (bend of elbow, back of knee and groin) and dry (volar forearm and palm) [4]. For example, sebaceous areas show the lowest bacterial diversity, and the Propionibacterium (including Cutibacterium) are the main isolates bacteria from these areas [2]. Different external factors, such as soaps, cosmetics, antibiotics, temperature, humidity, air pollution, and UV exposure could influence the microbiome composition [2,10-12]. Although efficacious, the treatments for acne could decrease the richness and evenness of the cutaneous microbiome [9]. Also the treatments not considered to have antimicrobial properties (e.g., isotretinoin), could alters the composition of the cutaneous microbiome [13]. In addition to systemic or topical treatments, skin cleansing protocols represent an important step in acne care [14]. Indeed, the used of suitable cleansers reduces the side effects of acne treatments and increased their therapeutic efficacy [15-17]. The suitable cleanser should be non-acnegenic, non-comedogenic, non-irritant and without allergens [17], indeed, the used of aggressive cleansers can be associated with an increased risk of inducing skin barrier impairment, dry skin and/or local dysbiosis. Therefore, the use of cleansing products that preserve or restore the microbiome balance represents a key strategy in the holistic treatment approach of acne.
Study aim
The aim of this study was to evaluate the skin acceptability and the effect on skin face microbiome of a cleanser containing Aloe Vera and Biopep15 (an oligopeptide with antibacterial and antiinflammatory activities) in subjects with acne-prone skin.
Population and study design
A 28 day prospective open label trial was conducted in subjects with acne-prone skin. The clinical trial, conducted by Eurofins Consmetic and Personal Care Italy S.r.l. CRO (Vimodrone, Italy), was performed between November 2022 and December 2022. The study was performed according to Good Clinical Practice Guidelines and the Declaration of Helsinki. All subjects provided signed informed consent prior to start the study. Seventeen subjects were enrolled in the study (12 female and 5 male, median age 23 years). Fifteen subjects (88%) complete the study phases. Two subjects stopped prematurely the trial due to personal reasons independent of the study. The main inclusion criteria were age between 18-40 years and greasy skin prone to acne. The main exclusion criteria were a personal history of adverse reaction to the product/ingredients used in the study, and the used of treatments able to interfere with the interpretation of the results. The study was conducted over 28 days, patients were evaluated at baseline and at the end of the study. Patients applied the cleanser (Biretix Cleanser, Cantabria Labs, Madrid, Spain) twice daily, morning and evening, to wet skin, massaging gently. The product was let act 1-2 minutes, then was rinse thoroughly.
Study outcomes
The main outcomes of this study were to evaluate the skin acceptability of cleanser and it effect on skin microbiome. The study foresaw 28 days of treatment; checks points were scheduled at baseline (T0/D0) and after 28 days (D28). The tolerability of cleanser was evaluated by the investigating dermatologist, in collaboration with participants, based on both physical (dryness, erythema, oedema, desquamation) and functional (burning sensation, itching sensation, tight feeling, other) signs. The effect of cleanser on microbiome was evaluated analysing bacteria genes present on the skin and their relative abundance at the inclusion (D0/T0) and then after 28 consecutive days of product use at home (D28). For specimen sampling to analyse the skin bacterial community, the study skin area was swabbed with sterile swabs. The genomic DNA from the swab heads was extracted using Ion 16S Metagenomics Kit (ThermoFisher Scientific). The Ion 16S™ Metagenomics Kit uses two primer pools to amplify seven hypervariable regions (V2, V3, V4, V6, V7, V8 and V9) of bacterial 16S rRNA. The amplified fragments can then be sequenced on either the Ion S5™ or the Ion PGM™ Systems, and analysed using the Ion16S metagenomics workflow in Ion Reporter™ Software. Ion Reporter Software enables the rapid identification (at genus or species level) of microbes present in complex polybacterial research samples, using both curated Greengenes and premium curated MicroSEQ ID 16S rRNA reference databases. For each experimental time, the relative abundance of each phylum detected at a level>1% was considered. Different indexes were calculated: Observed OTU (number of operational taxonomic units); Chao1 Index (“Chao1”, richness index taking into account rare OTUs); Shannon Diversity Index (“Shannon”, diversity index taking into consideration the abundance of each taxon and whether or not the distribution of OTUs in the sample is even) [18].
Statistical analysis
Statistical analyses were conducted on the skin microbiome data. Continuous variables were expressed as mean ± Standard Deviation (SD) and the variation in percentage versus baseline (D0/T0) at the end of treatment (D28) was calculated. The significance was calculated according to p-value of bilateral Student t test in pairs, in case of normality of the distribution or the Wilcoxon test in the adverse case. The variations are judged statistically significant if p<0.05. In view of the proof-of-concept nature of the present trial, a formal sample size calculation was not performed. We decided to enroll at least 15 evaluable subjects.
A total of 17 patients with a median age of 23 years were enrolled in the study (12 female and 5 male, median age 23 years). Fifteen subjects (88%) complete the study phases. Two subjects stopped prematurely the trial due to personal reasons independent of the study. The skin acceptability was evaluated considering both clinical signs (skin examination) and sensation of discomfort reported by the subjects. After 28 consecutive days of use of the investigational product at home, no clinical signs imputable to the investigational product were observed by the dermatologist. After questioning, no subject declared sensations of discomfort due to the application of the investigational product. The effect of cleanser on skin microbiome was evaluated applying the DNA sequencing techniques, that allowed to analyse specimens and account for all the bacterial genes present at a given time, as well as their relative proportion, before and after the cosmetic treatment. The relative abundance of each phyla and genera detected on T0 and after 28 days of treatment at level>1% was reported in Table 1.
| Phyla/genera | Time | Relative abundance | Δ (D28-D0) |
|---|---|---|---|
| (mean ± SD) | |||
| Actinobacteria | D0 | 53.52 ± 25.15 | -0.38 ± 23.04 |
| D28 | 53.14 ± 19.74 | ||
| Corynebacterium | D0 | 6.42 ± 5.22 | -1.86 ± 3.15* |
| D28 | 4.56 ± 4.06 | ||
| Propionibacterium | D0 | 38.13 ± 22.81 | 3.39 ± 21.6 |
| D28 | 41.52 ± 18.47 | ||
| Bacteroidetes | D0 | 1.07 ± 1.24 | 2.41 ± 4.72* |
| D28 | 3.48 ± 4.32 | ||
| Firmicutes | D0 | 29.36 ± 21.47 | -7.14 ± 17.06 |
| D28 | 22.22 ± 8.59 | ||
| Staphylococcus | D0 | 14.60 ± 17.51 | -1.50 ± 15.89 |
| D28 | 13.10 ± 9.26 | ||
| Streptococcus | D0 | 7.11 ± 12.58 | -2.93 ± 11.10 |
| D28 | 4.18 ± 3.64 | ||
| Proteobacteria | D0 | 14.74 ± 16.54 | 5.48 ± 17.75 |
| D28 | 20.22 ± 18.34 | ||
| Acinetobacter | D0 | 3.24 ± 6.31 | -0.53 ± 5.29 |
| D28 | 2.71 ± 2.56 |
Note: * p<0.05
Table 1: Relative abundance of phyla and genera (level>1%) detected on T0 and after 28 days of treatment (n=15).
At phylum level, members of four phyla were predominant: Actinobacteria (53.52 ± 25.15%), Bacteroidetes (1.07 ± 1.24%), Firmicutes (29.36 ± 21.47%), Proteobacteria (14.74 ± 16.54%). Within these phyla Corynebacterium, Propionibacterium, Staphylococcus, Streptococcus and Acinetobacter were the most abundant genera, accounting for 69.50% of the sequences at D0/T0 and 66.06% at D28. The analysis of the results reflecting the long-term effect of the product following 28 consecutive days of application showed a decrease in Actinobacteria (-0.38 ± 23.04%) and Firmicutes (-7.14 ± 17.06%), and an increase in Proteobacteria (5.48 ± 17.75%) and Bacteroidetes (2.41 ± 4.72%, p<0.05), although generally not statistically significant.
At genus level a statistical decrease of Corynebacterium (-1.86 ± 3.15%, p<0.05), a decreased of Staphylococcus and Streptococcus and Acinetobacter (-1.50 ± 15.89%, -2.93 ± 11.10%, -0.53 ± 5.29% respectively), and an increase of Propionibacterium (3.39 ± 21.6%) were observed at the end of the treatment.
After the qualitative and quantitative microbiome evaluation, different indexes were calculated. The Observed OTU represents the number of operational taxonomic units. Chao1 Index is a richness index, taking into account rare OTUs. Shannon Diversity Index is a diversity index, taking into consideration the abundance of each taxon and whether or not the distribution of OTUs in the sample is even [18]. The mean Chao1 index and the mean OTUs index, representing bacterial richness, and the mean Shannon index, a representative diversity index, were calculated over the whole population (Figure 1).
The use of the cleanser did not alter microbial richness and microbial diversity evaluated by Chao1, mean OTUs and Shannon Index. A slight increase, although not significant, in microbial diversity was observed after 28 days of treatment.
In recent years, an increasing attention has been given to the human skin microbiota and microbiome, since an imbalance in skin microbiome, is correlated with skin pathological diseases, such as acne. Negative effects on skin microbiome were described also after specific acne treatments [13]. Although, skin cleansing protocols represent an important step in acne care, aggressive cleanser could be associated with negative effects, such as local dysbiosis. To maintain or restore the microbial homeostasis represent an important strategy for the formulation of cosmetics products [19]. A cleaning product able to preserve or restore the microbiome balance could therefore represent an important tool in the treatment approach of acne.
In this context, a prospective open label trial was conducted to evaluate the skin acceptability and the effect of a cleanser containing Aloe Vera and Biopep15 on skin microbiome in subjects with acne-prone skin. Biopep15 is an oligopeptide with antibacterial activity that can interfere with lipoteichoic acid (a component of the wall of Cutibacterium acnes) [20]. Biopep 15 could also interfere with the Cutibacterium acnes TLR2 activation which is considered a crucial pro-inflammatory step in acne formation. In the present study the investigational product has shown a very good skin acceptability: no clinical signs or sensations of discomfort were observed at the end of the treatment (28 days). The effect on skin microbiome was evaluated by means of skin DNA sequencing, after 28 days of product used, calculating also different indexes (Chao1, OTUs and Shannon).
The importance of microbiome diversity and richness has recently been highlighted by research community because was observed an association between skin microbiome and dermatologic conditions (e.g. acne and atopic dermatitis) [21]. A loss of diversity and richness is generally observed in acne patients [9,22]. As reported in Figure 1, the use of the cleanser did not alter microbial richness and microbial diversity. In addition, a slight, although not significant, increase in microbial diversity was observed after 28 days of treatment.
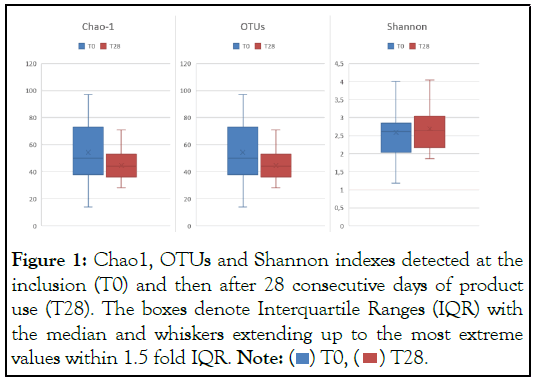
Figure 1: Chao1, OTUs and Shannon indexes detected at the
inclusion (T0) and then after 28 consecutive days of product
use (T28). The boxes denote Interquartile Ranges (IQR) with
the median and whiskers extending up to the most extreme
values within 1.5 fold IQR. Note:  .
.
The results reflecting the long-term effect of the tested cleanser showed a decrease in Actinobacteria and Firmicutes, and an increase in Proteobacteria, although generally not statistically significant. Actinobacteria and Firmicutes are generally increased in acne patients while Proteobacteria are usually underrepresented in the acne skin patients [7,9]. In addition, a significant increase in Bacteroidetes was observed at the end of treatment (from 1.07 to 3.48%, p<0.05), obtaining data more similar to the relative abundance of this phyla reported in the literature (6.3%) [9]. At genus level a statistical decrease of Corynebacterium and a decreased of Staphylococcus and Streptococcus (both generally abundant on acne skin), were observed [7].
Some limitations should be considered in evaluating our results. First, this is a proof-of-concept study. Future trials with larger sample size are required to evaluate the real clinical effects of this products to modulate the skin microbiome. A second limitation of this study is correlated with the difficulties associated to the methods used in microbiome research that required standardised and validated protocols and could be associated with bias, such as potential contaminations. However, compared to the in vitro methods, the in vivo studies allow a better characterization of microbiome and the evaluation of the possible role of cosmetics in its modulation.
The cleanser was applicated on the face twice a day (morning and evening) for 28 consecutive days. The skin acceptability (clinical signs and sensations of discomfort) and the effect on skin microbiome (by means of skin DNA sequencing) were evaluated after 28 days of product used. The cleanser tested in this study was well tolerated by all participants with preserving and restoring effects on microbiome balance in acne patients. Therefore, it could be used as adjuvant product for effective acne treatments like benzoyl peroxide and oral isotretinoin which could have negative effects on skin microbiome or as cleansing protocol to maintain the therapeutic results.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Colombo F, Milani M (2023) The Role of an Aloe Vera and Antimicrobial Peptide-Based Skin Cleanser on Microbiome Balance in Acne Vulgaris: A Prospective, Proof of Concept, Open Label Trial. J Clin Exp Dermatol Res. 14:632.
Received: 27-Feb-2023, Manuscript No. JCEDR-23-22949; Editor assigned: 01-Mar-2023, Pre QC No. JCEDR-23-22949; Reviewed: 15-Mar-2023, QC No. JCEDR-23-22949; Revised: 27-Mar-2023, Manuscript No. JCEDR-23-22949; Published: 31-Mar-2023 , DOI: 10.35841/2155-9554.23.14.632
Copyright: © 2023 Colombo F, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.