Journal of Cell Signaling
Open Access
ISSN: 2576-1471
ISSN: 2576-1471
Research Article - (2020)Volume 5, Issue 1
Ellagitanins are natural and complex polyphenolic compounds enriched in foods and commercial supplements. They are initially hydrolyzed to ellagic acid and further metabolized to urolithins by the gut microbiome. Ellagitanins are among a number of endogenous compounds found in fruits, nuts and vegetables that comprise a cardio protective diet for humans; however, biotransformation to urolithins may underlie their protective effects. Chronic treatment with urolithin A conveys anti-fibrotic and anti-inflammatory actions in experimental models of brain, cardiac, and kidney injury. Since the cellular actions and signaling events of urolithin A are undefined in renal cells, we assessed the influence of urolithin A on the TGF-β-PAI-1 pathway in NRK-52e cells, a well-characterized model of the proximal tubule epithelium and TGF-β induced signaling. TGF-β stimulated PAI-1 release 12-fold which was abolished by both urolithin A and the TGF-β receptor kinase inhibitor SB525334. The PAI-1 response to TGF-β was also blocked by the EGF receptor (EGFR) kinase inhibitors AG1478 and lapatinib implicating EGFR transactivation by TGF-β to evoke PAI-1 release in the renal cells. Indeed, EGF directly stimulated PAI-1 release that was abolished by urolithin A as well as the EGFR inhibitors. Moreover, both urolithin A and AG1478 blocked EGFR auto phosphorylation induced by EGF. The present studies suggest that the microbiome product urolithin A abrogates the stimulated release of PAI-1 by blocking the phosphorylation (activation) of the EGFR in NRK-52e renal epithelial cells.
Urolithin A; PAI-1; TGF- ; EGF; EGFR transactivation; NRK-52e cells
ADAMs: Membrane-anchored disintegrin metalloproteases; AKI: Acute Kidney Injury; AKT: Protein Kinase a; Ang II: Angiotensin II; CKD: Chronic Kidney Disease; COX2: Cyclooxygenase 2; EGF: Epidermal Growth Factor; EGFR: Epidermal Growth Factor Receptor; EMT: Epithelial Mesenchymal Transition; GPCRs: Receptors coupled to G proteins; LDH: Lactate Dehydrogenase; MAPK: Protein kinases activated by mitogen; MMPs: Metalloproteinases; NFKB: Nuclear factor-κB; OAT4: Organic acid transporter 4; PAI-1: Plasminogen activator inhibitor-1;PI3K: Phosphatidylinositol 3- kinase; PKC: Protein kinase C; PLC: Phospholipase C; ROS: Reactive Oxygen Species; SGLT1: Sodium-glucose cotransporter; SMAD: Mothers against decapentaplegic homolog; TGFβ: Transforming growth factor-β; TGFβR: Transforming Growth Factor-β Receptor; TLR4: Toll-Like Receptor 4; TNFα: Tumor Necrosis Factor-α; UAG: Urolithin A Glucuronide
Ellagitanins are natural and complex polyphenolic compounds enriched in certain fruits including red grapes, strawberries and pomegranates as well as in nuts and teas. They are also found in various commercial supplements, some of which are derived from extracts of the muscadine grape that are particularly high in polyphenolic content [1-4]. Ellagitanins are among a number of endogenous compounds in fruits, nuts and vegetables that comprise a cardioprotective diet for humans [2-4]. The initial acid-dependent hydrolysis of ellagitanins generates ellagic acid which serves as the precursor to the microbiome-dependent generation of urolithins in the gut [2-5]. Thus, urolithins are the end stage products of ellagitanin metabolism that are catalyzed in humans by the microbial bacteria Gordonibacter and Ellagibacter isourolithinfacines [5]. Numerous studies reveal that chronic treatment with ellagic acid or urolithin A convey antifibrotic and anti-inflammatory properties in rodent models of brain, cardiac, kidney and vascular injury [6-19]. The beneficial effects of urolithin A suggests that the microbiome-dependent metabolism to urolithin A may be critical to the functional properties of ellagitanins or ellagic acid which, in part, may reflect the increased bioavailability of urolithins [1,3,5,14]. Indeed, Guada et al. [10] report that urolithin A was more potent than ellagic acid in protecting the kidney from cisplatininduced tubular injury. Renal indices of damage including tubular degeneration and apoptosis, elevated serum creatinine levels and increased expression of inflammatory cytokines were significantly reduced by urolithin A treatment in the cisplatin injury model [10]. However, the signaling mechanisms of urolithins that potentially contribute to their influence on fibrotic and inflammatory pathways within the kidney are unknown.
Giménez-Bastida et al. provided initial evidence that urolithin A reduced plasminogen activator inhibitor-1 (PAI-1) release, a key promotor of tissue fibrosis, in colonic fibroblast cells exposed to the cytokine IL-1β [20]. PAI-1 is a major downstream mediator of both transforming growth factor-beta (TGF-β) and epidermal growth factor (EGF) pathways within the kidney that contribute to fibrosis, inflammation, and compromised renal function and ultimately end stage renal failure [21-30]. PAI-1 blocks the conversion of plasminogen to plasmin and the subsequent activation of fibrolytic pathways to mitigate fibrosis [27]. PAI-1 knockout mice or PAI-1 inhibitor-treated animals have an attenuated injury response in experimental models of renal inflammation and fibrosis [27,28,31-33]. PAI-1 may also contribute to various pathological events including hypertension, cellular senescence, myofibroblast transition, TGF-β stimulation, autophagy, and oxidative stress [26-29,33-37]. Since the cellular actions of urolithin A have not been established in renal epithelial cells, we determined whether urolithin A attenuates the PAI-1 pathway in the rat kidney NRK-52e cell line, a well-characterized proximal tubule model for both TGF-β and EGF-induced fibrotic pathways [38-45]. The tubular epithelium is a key site in the progression of renal injury, particularly for the deleterious effects of TGF-β and EGF [24,26,46-51]. Moreover, the extent of tubulointerstial fibrosis is a major predictor of chronic kidney disease and renal failure [24,48-51].
The current study demonstrates that the TGF-β evoked increase in the cellular release of PAI-1 (~12 fold) was abolished by urolithin A and the TGF-β receptor (TGβR) kinase inhibitor SB525334 in NRK-52 cells. The stimulated release of PAI-1 by TGF-β was also blocked by the EGF receptor (EGFR) kinase inhibitors AG1478 and lapatinib implicating the transactivation of EGFR in the PAI-1 response through a liganddependent pathway. Indeed, EGF directly stimulated the release of PAI-1 which was abolished by urolithin A, as well as AG1478 and lapatinib. Moreover, both urolithin A and AG1478 abolished the EGF-dependent phosphorylation of EGFR suggesting a similar mode of inhibition for these two compounds. The present findings demonstrate that urolithin A abolished the stimulated release of PAI-1 that may reflect the inhibition of EGFR phosphorylation (activation) in NRK-52e renal cells.
Cell culture
Normal kidney proximal epithelial cells (NRK-52e) cells were obtained at passage 15 from American Tissue Type Culture (ATTC, Arlington VA). Cells were maintained at 37°C in plastic 75 cm2 flasks in Dulbecco’s modified Eagle’s medium (DMEM) containing 5% calf serum, 4.5 gm/L glucose, 2 mM LGlutamine and 1.5 gm/L bicarbonate, but without antibiotics including penicillin and streptomycin per ATTC recommendations. Following passage into 24-well plates or 100 mm dishes, cells near confluence were washed in Dulbecco’s phosphate buffered saline (PBS) to remove the serum and maintained in serum-free DMEM for 48 h prior to TGF-β or EGF treatment; the cell media was replaced daily. Cells of passage 18 to 30 were used in these studies and each experiment reflects a different cell passage. Cell toxicity to urolithin A in the presence or absence of TGF-β was assessed by the release of lactate dehydrogenase (LDH) in the cell media using a LDH cytotoxicity assay kit (Thermo Scientific, Rockford, IL USA).
Cell treatments
Cells incubated in serum-free media for 48 h were replenished with fresh media and pre-treated with urolithin A or other inhibitors 2 h prior to the stimulation of PAI-1 release with TGF-β or EGF (5 ng/mL each, Peprotech, NJ, USA) for 24 h. Blockade of EGFR phosphorylation was determined by prior treatment with AG1478 (1 μM) or urolithin A (50 μM) followed by a 5 min exposure to EGF (5 ng/mL). Total EGFR expression was assessed in cells following a 24 h exposure to 50 μM urolithin A. Urolithin A was obtained from Cayman Chemical (Ann Arbor MI, USA); EGFR inhibitors AG1478 and lapatinib, TGβR kinase inhibitor SB525334 and the PKC inhibitor Go6983 were from Selleckchem (Houston, TX, USA); the cSRC kinase inhibitor SU6656 from APExBio (Houston TX, USA); and the metalloproteinase/ADAM inhibitor GM6001 from MedChem Express (Monmouth Junction NJ, USA). Agents were initially dissolved in 100% DMSO and stored in stock solutions at -80ºC. The maximal concentration of DMSO in the cell studies was 0.2% vol/vol which was tested alone on PAI-1 release.
PAI-1 measurement
PAI-1 release was quantified in the cell media from duplicate wells (0.5 ml each) of 24-well plates using a PAI-1 (Serpine 1) SimpleStep ELISA (ab201283, Abcam) from cells of separate passages. Cell samples were routinely batched from different experiments and assayed with the same ELISA kit. The cell media samples were diluted at least 1:1 (vol/vol) in the ELISA buffer and directly added to the 96-well assay plate. PAI-1 in the DMEM media alone was undetectable and the cell media did not interfere or quench PAI-1 immunoreactivity using the PAI-1 standards (data not shown). The sensitivity of the assay was 0.16 nanograms per mL (ng/mL) based on the ELISA specifications and the PAI-1 release expressed as ng/mL of cell media. For some studies, the inhibitory response was expressed as a percentage (%) of the TGF-β-PAI-1 release set to 100%.
EGF receptor phosphorylation and expression
EGF receptor (EGFR) phosphorylation and EGFR expression were determined by immunoblotting. Cells in 100 mm dishes were scraped into 2 mL microcentrifuge tubes with ice cold PBS and pelleted at 5,000 g for 3 min at 4°C; the buffer was removed and the cell pellets were immediately frozen at -20°C. Cell pellets were prepared in 0.1 mL PBS containing orthovanadate (1 mM), sodium fluoride (50 mM), EDTA (5 mM), PMSF (0.1 mM) and a Sigma Protease Inhibitor cocktail (10 μL/mL; P8340) and immediately sonicated at an amplitude of 30% (Ultrasonic processor, Thermo-Fisher Scientific, Chicago, IL). Protein was quantified using a BioRad Protein kit. The cell homogenate was diluted with an equal volume of Laemmli buffer and boiled for 5 minutes. Samples of 25 micrograms (μg) were loaded onto a BioRad TGX Stain free 10% gel and proteins were separated electrophoretically at 120 volts for 70 min followed by transfer to a PVDF membrane using a BioRad Trans-Blot TurboTM transfer system. Membranes were blocked with 5% milk for 60 min at RT and probed overnight at 4°C with primary antibodies for the EGFR (1:1000, C74B9, Cell Signaling, Cambridge MA) or phospho-EGFR (1:1000; Tyr-1068, D7A5, Cell Signaling). The following day, membranes were incubated for 60 min with an anti-rabbit secondary antibody (1:5000, NA934, GE Healthcare, Boston MA) in 5% milk at RT followed by a 5 min incubation with Femto (1:5 dilution, Thermo Scientific); reactive bands were visualized with the BioRad ChemiDoc system. Membranes probed with the phospho-EGFR membranes were stripped, re-probed for total EGFR and expressed as the ratio of phospho-EGFR to total EGFR. EGFR was expressed as the ratio of EGFR to the total protein in each lane.
Statistical analysis
The data were expressed as mean ± standard error (SEM). Differences between multiple groups were assessed by one-way ANOVA (Tukey’s post-hoc test) and differences between two groups were assessed by a two-tailed t-test using the GraphPad Prism 6.2 statistical and plotting software (San Diego, CA USA) (Figure 1).
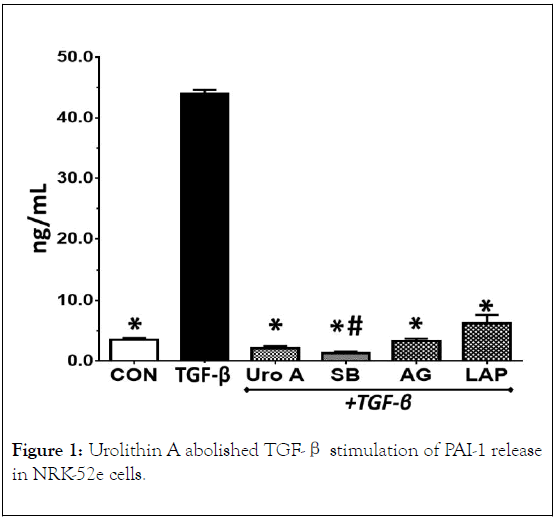
Figure 1. Urolithin A abolished TGF-β stimulation of PAI-1 release in NRK-52e cells.
TGF-β (5 ng/mL) stimulated PAI-1 release approximately 12- fold over control cells (CON) after 24 h. Urolithin A (Uro A, 50 μM) and the TGF-β receptor kinase inhibitor SB525334 (SB, 1 μM) abolished the increase in PAI-1 release by TGF-β. The EGF receptor kinase inhibitors AG1478 (AG, 1 μM) and lapatinib (LAP, 1 μM) essentially abolished PAI-1 release. Data are the means ± SEM from 3 separate experiments; *p< 0.01 vs. TGF-β, #p<0.05 vs.CON.
Urolithin a abolishes tgf-β stimulation of pai-1 release
The basal release of PAI-1 ranged from 3 to 5 ng/mL over a 24 h period which was comparable to basal PAI-1 levels previously reported in NRK-52e cells (Figures 1-4) [37]. A 24 h treatment with TGF-β (5 ng/mL) increased PAI-1 release approximately 12-fold (3.8 ± 0.2 vs. 44 ± 3.0 ng/mL, P<0.05). Pretreatment of the cells with the TGF-β receptor (TGβR) kinase inhibitor SB525334 (1 μM) abolished the PAI-1 response to TGF-β; urolithin A pretreatment (50 μM) also abolished the cellular release of PAI-1. Urolithin A (Uro A, 50 μM) significantly reduced LDH release into the cell media in the presence of TGF-β (5 ng/mL) (A) or alone (B) after a 24 h exposure. Data are the mean ± SEM from 3-4 separate experiments; *p< 0.05 (A) and *p<0.02 (B).
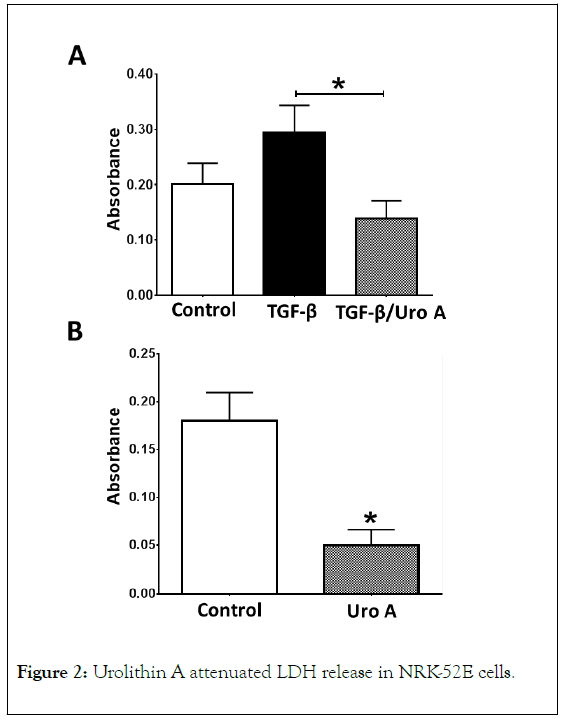
Figure 2. Urolithin A attenuated LDH release in NRK-52E cells.
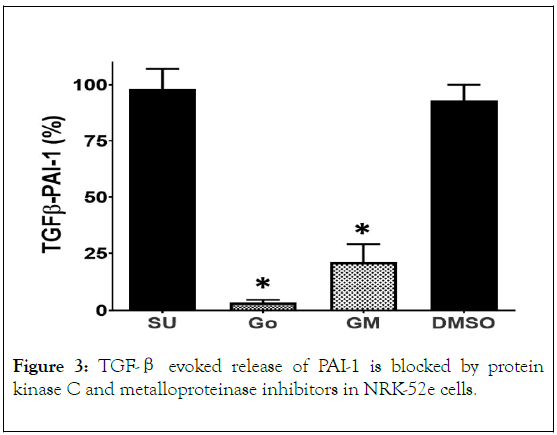
Figure 3. TGF-β evoked release of PAI-1 is blocked by protein kinase C and metalloproteinase inhibitors in NRK-52e cells.
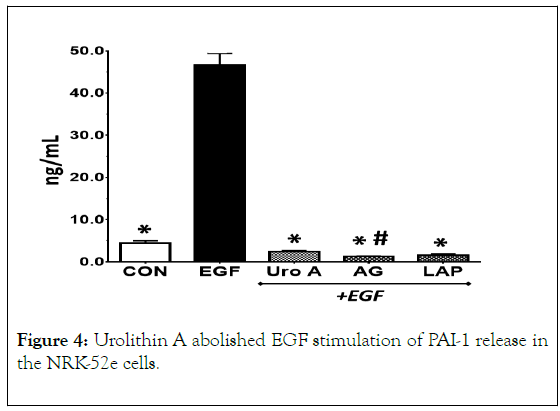
Figure 4. Urolithin A abolished EGF stimulation of PAI-1 release in the NRK-52e cells.
The effects of urolithin A (50 μM) on cell toxicity were assessed by measuring LDH release over a 24 h incubation period. As shown in Figure 2A, urolithin A significantly reduced LDH release in the presence of TGF-β. In separate studies, urolithin A (50 μM) also significantly reduced LDH release in the control cells after 24 h (Figure 2B).
The cSRC kinase inhibitor SU6566 (SU, 10 μM) and DMSO alone (0.2%) failed to block PAI-1 release to TGF-β (5 ng/mL). The protein kinase C inhibitor Go6983 (Go, 10 μM) and the metalloproteinase inhibitor GM6001 (GM, 10 μM) significantly blocked PAI-1 release. Data are expressed as % of the TGF-β evoked PAI-1 response and are the means ± SEM of 3 separate experiments; *p< 0.05 vs. TGF-β.
TGF-β transactivates EGFR
TGF-β is reported to stimulate PAI-1 release by activation of the EGF receptor (EGFR transactivation) in various cell types [21,23,25,29,30,45,52,53]. Thus, we assessed the role of the EGFR in mediating the effects of TGF-β on PAI-1 release in the renal cells. As shown in Figure 1, pretreatment of the NRK-52e cells with the EGFR kinase inhibitor AG1478 or the EGFR/HER2 kinase inhibitor lapatinib essentially abolished the release of PAI-1 by TGF-β. Transactivation of EGFR by TGF-β may occur through a ligand-independent process involving the phosphorylation of the receptor by cSRC kinase [21-30,54-56]; however, pretreatment of the cells with the selective cSRC inhibitor SU6566 (SU) failed to attenuate the PAI-1 response to TGF-β (Figure 3). Alternatively, TGF-β through the activation of protein kinase C (PKC) stimulates metalloproteinases including matrix metalloproteinases (MMPs) and ADAMs that can release or shed EGF from the cell membrane [23,29,30,44,52-59]. As shown in Figure 3, the pan PKC inhibitor Go6983 (Go) abolished PAI-1 release while the metalloproteinase inhibitor GM6001 (GM) significantly inhibited the release of PAI-1 by TGF-β. Also note that the maximal concentration of DMSO alone (0.2%) did not significantly inhibit PAI-1 release (Figure 3).
EGF (5 ng/mL) stimulated PAI-1 release as compared to control cells (CON) after 24 h. Urolithin A (Uro A, 50 μM) and the EGF receptor kinase inhibitors AG1478 (AG, 1 μM) and lapatinib (LAP, 1 μM) abolished PAI-1 release. Data are the mean ± SEM from 3 separate experiments; *p< 0.01 vs. EGF; #p<0.05 vs. CON.
Urolithin a abolishes EGF stimulation of PAI-1
We examined EGF stimulation of PAI-1 release and attenuation of this response by urolithin A. EGF (5 mg/mL) also stimulated PAI-1 release in NRK-52e cells which was abolished by the EGFR kinase inhibitors AG1478 and lapatinib, as well as by urolithin A (Figure 4).
EGF (5 mg/mL, 5 min) stimulated phosphorylation of the EGF receptor (pEGFR, 170 kDa). AG1478 (AG, 1 μM) and urolithin A (Uro A, 50 μM) abolished EGFR phosphorylation. Blots shown are 2 representative experiments (A, B). Panel C are the means ± SEM of the pEGFR: EGFR ratio from 4 separate experiments; *p< 0.05.
We next determined whether urolithin A blocks EGF-stimulated phosphorylation of the EGFR. As shown in Figure 5, acute EGF exposure (5 min) increased EGFR phosphorylation approximately 4-fold as demonstrated by the increased immunoreactivity to the consensus autophosphorylation site (Tyr-1068) of the EGFR. Pretreatment with the EGFR kinase inhibitor AG1478 or urolithin A abolished EGFR phosphorylation by EGF (Figures 5A-5C).
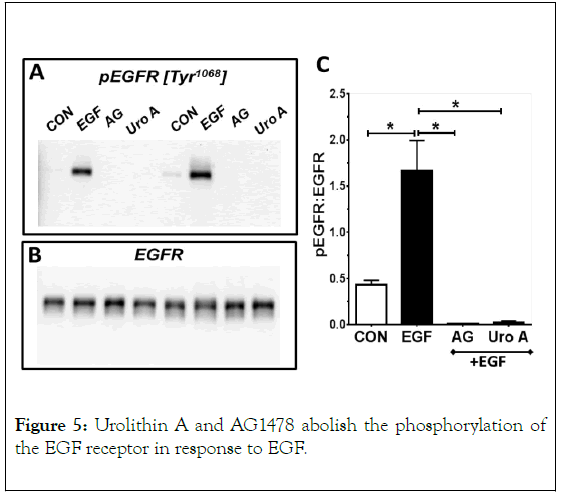
Figure 5. Urolithin A and AG1478 abolish the phosphorylation of the EGF receptor in response to EGF.
Urolithin A (Uro A, 50 μM) treatment of control cells for 24 h reduced the expression of the EGF receptor (EGFR, 170 kDa) by 32% (A, C). Data are ratio of EGFR (A) to total protein (B) and are the means ± SEM from 4 separate experiments; *p< 0.05.
Finally, we determined the effects of chronic urolithin A treatment (50 μM) on the expression of the EGFR for 24 h. As shown in Figure 6, urolithin A had a small, but significant, inhibitory effect on EGFR expression in comparison to the control cells. Urolithin A treatment reduced EGFR protein by 32% in NRK-52e cells in the absence of exogenous TGF-β or EGF.
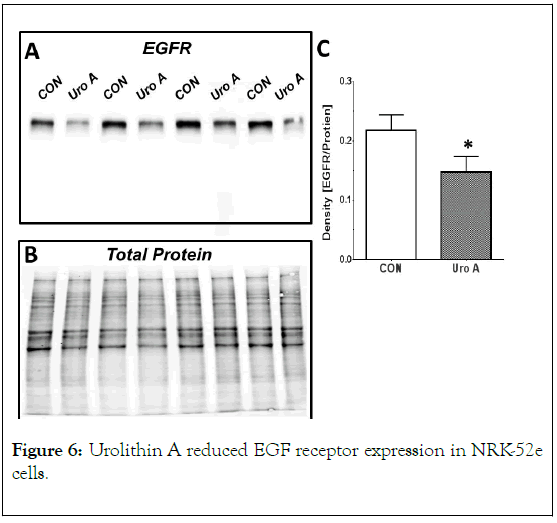
Figure 6. Urolithin A reduced EGF receptor expression in NRK-52e cells.
The present study demonstrates that the microbiome product urolithin A abolished the stimulated release of PAI-1 by TGF-β in NRK-52e renal epithelial cells. We show that the TGF-β- induced release of PAI-1 is dependent on transactivation of the EGFR in the renal cells and that urolithin A also abolished the PAI-1 response to EGF. Moreover, urolithin A abolished the phosphorylation of the EGFR in response to exogenous EGF, as well as attenuated EGFR expression. Urolithin A reduced the increase in LDH release by TGF-β suggesting that urolithin A lacks toxic actions and may exhibit overall protective effects in NRK-52e cells. The current findings strengthen the concept that the metabolism of dietary ellagitanins to urolithin A by the microbiome may result in generation of an endogenous compound with anti-fibrotic properties. Indeed, the current results in NRK-52e proximal tubule cells support recent studies demonstrating that the administration of urolithin A conveys protective effects in various models of brain, heart, vascular and kidney injury [6-19]. To our knowledge, the present study is the first to demonstrate that urolithin A mitigates the activation of the TGF-β/EGFR/PAI-1 pathway in renal epithelial cells.
Increased expression and release of PAI-1 by TGF-β plays a key role in the progression of fibrosis in the kidney [21-30]. PAI-1 expression is also associated with a greater incidence of cardiovascular events, atherosclerosis, vascular stiffness and hypertension [58-64]. In addition to blocking the activation of plasmin that directly attenuates the fibrolytic pathway, PAI-1 also promotes cell senescence, activates the TLR-4 receptor involved in innate immunity and may further induce TGF-β and myofibroblast transition (EMT) [26,27,28,33-38]. The TGF-β- dependent stimulation of PAI-1 may involve transactivation of the EGFR through two distinct cellular pathways. In the ligandindependent pathway, TGF-β stimulates increased oxidative stress and activation of cSRC kinase to directly phosphorylate the EGFR [21,23,28,29,30,34,53,55,56]. In contrast, the liganddependent pathway reflects PKC activation of metalloproteinases such as ADAMs to hydrolyze the pro-EGF ligand to EGF and subsequent stimulation of EGFR phosphorylation by EGF [23,29,30,44,52,55-59]. Both the EGFR kinase inhibitors AG1478 and lapatinib blocked TGF-β induced PAI-1 release in the proximal tubule cells. The PKC inhibitor Go6983 and the MMP/ADAM inhibitor GM6001 abolished or significantly reduced the PAI-1 response to TGF-β while the cSRC inhibitor SU6656 failed to attenuate PAI-1 release. Thus, the present findings suggest an obligatory role of EGFR transactivation in the TGF-β-PAI-1 response which may arise from the stimulation of PKC coupled to the MMP/ADAMdependent shedding of the EGF ligand to bind and activate EGFR (Figure 7).
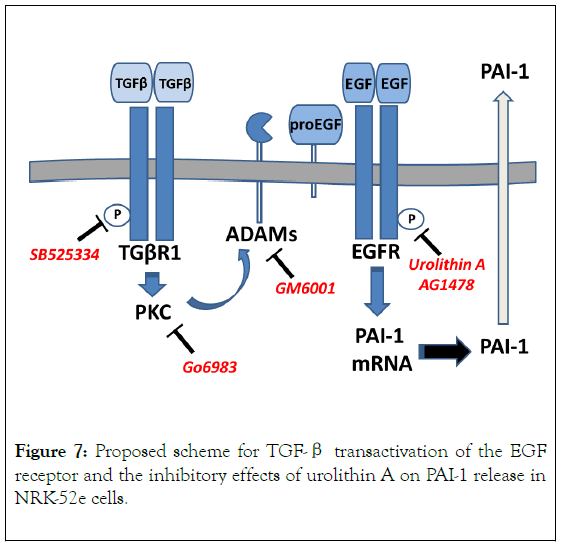
Figure 7. Proposed scheme for TGF-β transactivation of the EGF receptor and the inhibitory effects of urolithin A on PAI-1 release in NRK-52e cells.
Urolithin A abolished both the TGF-β and EGF stimulation of PAI-1 release in the NRK-52e cells. Urolithin A also abolished the phosphorylation of EGFR at the consensus autophosphorylation site on the receptor (Tyr-1048) following exposure to EGF, as well as reduced total EGFR protein expression after a 24 h treatment in control cells. The attenuation of EGFR phosphorylation by urolithin A was comparable to the EGFR kinase inhibitor AG1478 which may portend for a similar mechanism to inhibit the activation (phosphorylation) of EGFR. AG1478 binds to the ATP site of the tyrosine kinase domain of EGFR with an IC50 of 3 nM to block phosphorylation, but exhibits higher IC50 values in the 100 nM range to attenuate downstream effects in cell assays [23,55,65]. The potency of urolithin A to attenuate PAI-1 release is likely less than AG1478 which could reflect, in part, a reduced ability of urolithin A to access the intracellular compartment and/or a greater susceptibility to cellular metabolism as compared to the two tyrosine kinase inhibitors. Ellagic acid is reportedly internalized by the organic acid transporter (OAT4) and the sodium-glucose co-transporter (SGLT1) in human intestinal cells, but the mechanism and extent of urolithin A uptake in renal epithelial cells are unknown [66,67]. In regards to the cellular actions of urolithin A, PAI-1 undergoes constitutive secretion and the reduction in the extracellular levels of PAI-1 likely reflects inhibition of PAI-1 synthesis by urolithin A rather than impacting secretion of the protein [27]. Indeed, others show that the EGFR inhibitor AG1478 abolished the increase in PAI-1 mRNA levels and protein expression in response to TGF-β or EGF [28,45]. We acknowledge that additional studies are required to establish the potency and cellular handling of urolithin A, as well as the potential inhibition of other pro-fibrotic signaling events evoked by TGF- β including the effects on PAI-1 mRNA levels and protein expression in the renal tubule cells.
As to the translational benefit of urolithin A, the circulating levels of urolithin A following ingestion of an enriched dietary source of ellagitanins peak at 0.10 μM after several hours while local levels within the gut approach 40 μM [10,20]. However, the tissue levels of urolithin A within the kidney are unknown following an enriched diet or chronic treatment with urolithin A and ellagic acid. Moreover, urolithin A is glucuronidated to urolithin A glucuronide (UAG) which is readily filtered and excreted in the urine [3,5]. The urinary levels of UAG are reported to range from 1 to 10 μM and UAG may directly access the apical aspects of the renal tubular system [3,5]. Giménez- Bastida et al. reported that UAG but not urolithin A inhibited PAI-1 release induced by TNF-α in human endothelial cells [68]. However, urolithin A significantly reduced PAI-1 release by IL-1β in colonic fibroblasts [20]. The mechanism for the inhibitory effects of urolithin A and UAG on PAI-1 release, as well as the discrepancy between the two compounds to inhibit TNF-α-induced PAI-1 release were not addressed in the two studies. Various cell types or different stimuli and their associated signaling pathways may explain the contrasting responses to urolithin A and UAG. In preliminary studies, we also find that urolithin A abolished PAI-1 release to TGF-β in human HK-2 renal epithelial cells, an accepted model of human proximal tubules and fibrotic events (Chappell, unpublished observations). Currently, there are no commercial sources of UAG and the extent to which UAG attenuates PAI-1 release by TGF-β or EGF in either rat or human renal tubule cells awaits further study [69-71].
TGF-β induces TGF-β receptor 1 (TGβR1) autophosphorylation (P) and activates protein kinase C (PKC) to stimulate metalloproteinases such as ADAMs that cleave pro- EGF from the cell membrane. Soluble EGF stimulates EGF receptor (EGFR) autophosphorylation and downstream signaling events to augment PAI-1 mRNA levels and protein synthesis leading to an increase in the constitutive release of PAI-1. Urolithin A inhibits EGFR activation by TGF-β or EGF to attenuate PAI-1 release.
In conclusion, we demonstrate that the microbiome product urolithin A abolished both TGF-β and EGF-stimulated release of the pro-fibrotic protein PAI-1 in NRK-52e renal epithelial cells. The TGF-β-dependent stimulation of PAI-1 appears contingent on transactivation of the EGFR through a liganddependent pathway. Urolithin A blocked the EGF-dependent stimulation (phosphorylation) of the EGFR which may constitute one mechanism by which urolithin A attenuated the release of PAI-1 by TGF-β and EGF. In this regard, the protective effects of ellagic acid and urolithin A that are evident in various experimental models of tissue injury may reflect the functional actions of urolithin A to attenuate the TGF-β/ EGFR/PAI-1 pathway.
The authors gratefully acknowledge the support of the Wake Forest Baptist Comprehensive Cancer Center Cell Engineering Shared Resource, supported by the National Cancer Institute’s Cancer Center Support Grant award number P30CA012197. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute.
Funding for the current studies was provided by the Chronic Disease Research Fund, the Farley Hudson Foundation, the Hypertension & Vascular Research Center, the National Institutes of Health (NIH) R25HD047584 and R01HL146818, the American Heart Association (AHA) AHA-TPA34170522 and AHA-GIA151521.
None declared.
Mark Chappell: Conceptualization, Funding, Writing – Original draft preparation, Writing – Review & Editing, Visualization, Supervision, Methodology, Formal analysis, Investigation. Nancy Pirro: Methodology, Writing - Original draft preparation, Data curation, Visualization, Investigation; Ana Melo: Methodology, Visualization, Investigation. Ann Tallant: Writing – Review & Editing, Funding, Project Administration. Patricia Gallagher - Writing – Review & Editing, Funding, Project Administration.
Citation: Chappell MC, Pirro NT, Melo AC, Tallant EN, Gallagher PE (2020) The Microbiome Product Urolithin A Abrogates the TGF-β-EGFR-PAI-1 Pathway in NRK-52e Renal Epithelial Cells. J cell signal. 5:204. DOI:10.35248/2576-1471.20.5.204
Received: 31-Jan-2020 Accepted: 14-Feb-2020 Published: 21-Feb-2020 , DOI: 10.35248/2576-1471.20.5.204
Copyright: © 2020 Chappell MC, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Sources of funding : Funding for the current studies was provided by the Chronic Disease Research Fund, the Farley Hudson Foundation, the Hypertension & Vascular Research Center, the National Institutes of Health (NIH) R25HD047584 and R01HL146818,the American Heart Association (AHA) AHA-TPA34170522 and AHA-GIA151521.