Journal of Antivirals & Antiretrovirals
Open Access
ISSN: 1948-5964
ISSN: 1948-5964
Review Article - (2021)
The ongoing ravaging pandemic caused by SARS-CoV-2 continues to emerge by acquiring new mutations. Several recent mutations under different clades render the virus to bind more efficiently to the human ACE-2 (Angiotensin Converting Enzyme-2) with its spike protein. Detection inefficiency due to mutation in the spike protein (deletion of Δ69-Δ70 amino acid residues) misleads the flawless result of the COVID-19 test is also a significant concern. Several vaccine candidates have been designed to diminish this pandemic, from which four have been authorized due to exigency and more candidates are in the pipeline. The prime revealing mutations from different lineages (lineage B.1.177, B.1.1.7, B.1.351, and B.1.1.28 by PANGOLIN) has up risen several questions, including the vaccine efficacy against the mutated strains, alternate detection methods, evading potential of SARS-CoV-2 and clade lineage as well. The new variants recent incarnation has proved to have high transmissibility rather than the previous D614G variant and also indicates the latest outbreak of the mutated strain in an uncontrollable state. This review study aims to reveal these mutations along with discussing several vaccine strategies. The study also aims to describe several alternate detection methods to identify their clade and lineages and try to answer the arising questions from the current situation.
SARS-CoV-2; Clades; ACE-2; Vaccine; Lineage
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS- CoV-2) is a single-stranded positive-sense Ribonucleic Acid (RNA) virus that has burst upon the world, causing millions of deaths worldwide. Inadequate public health measures cause the virus to evolve more rapidly, acquiring several mutations [1]. Though the virus has a “Proofreading” activity, several lineages and clades have been established based on the characteristic given mutation [2- 4]. The natural mutation most probably occurs due to a chance event called the "Founder effect" that either helps the virus survive or be culled out from the population [5]. Since March 2020, the S:pD614G variant was predominant worldwide [1]. But in late December 2020, several new variants in the different geographical locations have emerged as a potential threat with increased transmissibility questioning the efficacy of the ongoing vaccination program [6]. The new European variants 20A.EU1 and 20A.EU2 with the characteristics mutations at S:pA222V and S:pS477N, including mutations in the nucleocapsid protein, has emerged as a potential threat [7]. The UK variants from the B.1.1.7 lineage with S:pN501Y mutations in the RBD of the spike protein, including the significant S:pN501Y, it has around 17 mutations with amino acid deletions Δ69-Δ70. The proportion of sequences has increased in the GISAID Epilfu database from 0.1% to 49.7% from early October to late November [8]. Along with the S:pN501Y mutations, the South African variants have emerged with other two crucial characteristics mutation S:pK417N and S:pE484K accounts for lower binding affinity with the monoclonal antibody [9]. The Brazilian variant has been grouped into two novel lineage P1 (S:pK417T and S:pE484K) and P2 (S:pN501Y) because of its significant divergence. However, S gene-based detection has been proved inefficient in detecting these mutating variants proposing alternative detection methods [10]. The current pandemic’s complete eradication of the virus largely depends on several vaccine strategies of which four are already authorized to introduce, and more are in the pipeline [11]. Several recent studies have shown that SARS-CoV-2 spike variant N439K can evade host antibody and other mutating strains, making it more worrying about the efficacy of the vaccination [12]. This current study summarizes the answers to the emerging questions raised by simultaneous mutation along with different clades and mutations, alternative detection of the virus, vaccine strategies, and several evasion mechanisms of the virus.
The SARS-CoV-2 virus is classified into seven major clades based on its marker mutation, namely S and L, later splitting L into G and V, and then G into GH, GR, and GV [13]. The S clade strains refer by nucleotide changes at positions C8782T and T28144C (NS8: Sp.L84S). The L clade strain is the wild version of the genome with no mutations. The V clade was designated by clustered co-occurring mutations at G11083T (NSP6: Sp.L37F) and G26144T (ORF3a: Sp.G251V). The mutation in nucleotides at C241T, C3037T, A23403G, and the amino acid change S-D614G was referred to as G clade. Additional G28882A mutation with amino acid changes in the N protein N:pG204R and ORF3a:pQ57H (G25563T) refer to GR and GH clade, respectively [2,14]. An additional mutation in the spike protein along with the S:pD614G, the nutation at position C22227T (S:pA222V), refers to the newly emerged GV clade [13].
The S:pD614G variant of SARS-CoV-2 became predominant and globally spread across different geographical regions from early March 2020. In the late summer of 2020, two mutating variants were prevailing in Spain, namely 20A.EU1 and 20A. EU2 (Nextstrain clade 20A) from the lineage of B.1.177. The 20A.EU1 has characteristics S:pA222V in the RBD of the spike protein, whereas the 20A.EU2 has S:pS477N amino acid substitutions, including mutations in the nucleocapsid protein [7]. In late December, due to the robust increase of COVID-19 patients in east and southeast England including London, surmise a new variant [4]. Complete analysis of the virus's genome was marked by 14 non-synonymous, four deletions, and six synonymous mutations. This also reveals several amino acid changes include deletion of amino acid residue at positions 69-70, deletion at positions 145, S:pN501Y, S:pA570D, S:pD614G, S:pP681H, S:pT716I, S:pS982A, and S:pD1118H [8]. This variant was named VOI 202012/1, also known as 501Y.V2 belonging to the GR clade (Nextstrain clade 20B and form B.1.1.7 lineage by PANGOLIN). The deletion at position S:pN501Y corresponds to a significant change in the RBD of the spike protein attributing to binding to ACE-2, whereas S:pP681H accounts for a change in the furin cleavage site [14]. A new variant was also identified spontaneously circulating in South Africa caused the second wave of COVID-19 in December 2020. Analysis of the virus grouped it in GH clades and B.1.351 lineage with amino acid changes in the spike protein and other regions in the cluster named 501Y.V2 [5]. These changes include S:pD80A, S:pD215G, S:pE484K, S:pA701V, along with the major S:pN501Y mutation. The three mutations in the spike receptor binding site (S:pK417N, S:pE484K and S:pN501Y) may increase receptor binding, affecting the binding of an antibody. Including these three mutations, 27% of GH clades' virus also contains nine major nucleotide deletions in NSP6 [15]. ‘Updated rapid risk assessment from ECDC on the risk related to the spread of new SARS-CoV-2 variants of concern in the EU/EEA-first update’, 2021). In January 2021, a new variant emerged in japan from travelers who have recently been to Brazil. Along with the new variant, another variant of the same lineage was also identified in Brazil. The first GR clade variant (Nextstrain clade 20B) belongs to the novel lineage P.1 (B.1.1.28) because of its divergence and have significant changes in the amino acid (S:pL18F, S:pT20N, S:pP26S, S:pD138Y, S:pR190S, S:pK417T, S:pE484K, S:pN501Y, S:pH655Y, S:pT1027I, and S:pV1176F) of its spike protein. The latter was termed as P2 with the S:pE484K mutation. The newly emerged UK variant, acquiring 17 mutations including three in-frame deletions and deletion of 69-70 amino acid residues, implicates a major change in the spike protein which sometimes causes failure of detection in the RT-PCR reactions based on S gene producing a false negative result) [10,16]. The S:pN501Y mutation is important as it enhanced the binding of spike protein to ACE-2 more tightly.
Several mathematical modeling studies mainly determine transmission probability. The European variant although does not account for a second wave but its transmissibility arose due to the lower restriction in traveling. And after Europe, it became the dominant variant in America indicating a higher transmission rate [7]. Early reports showed that the new variant VOC 202012/1 is 75% more transmissible than the other circulating variants with a R0 estimation of 0.39-0.93 for SARS-CoV-2 [17]. However, R values generated using different mathematical models based on mutational characteristics also propose a higher transmissibility rate. The South African variant 501Y.V2 also has a higher transmissible rate (56%) than the three main circulating variants three main variants (B.1.1.54, B.1.1.56, and C.1) in South Africa [4]. But no data was found for the diverge novel P.1 lineages: but with similar mutations, it may show the same transmission rate [16].
Several methods have been developed to detect SARS-CoV-2. Most of the tests are based on either nucleic acid testing or serological detection. However, Computed Tomography (CT scan) is also used nowadays. High throughput PCR-based detection is a detection method for the presence or absence of viral RNA. In this method, NP and OP swab samples are collected in Viral Transport Media (VTM), and subsequent RNA extraction is performed. cDNA synthesis is performed in 96 well plates and PCR is performed using ten picomoles of each forward and reverse primer. After that, capillary electrophoresis is performed to identify positive samples in each PCR product. Three or more than three gene primer pairs are used. This is an efficient method to achieve a high throughput result and it could reduce dependency on RT-PCR [18]. Immunoassays are also available for detection of SARS-CoV-2 also called point of care lateral flow test. IgM is generated in the host's body after 3-6 days of infection, while IgG is produced after eight days of infection. These antibodies can be detected in the blood of a patient. In this method, 10-15 μl of blood is added to a sample port containing 70 μl of sample dilution buffer. This blood specimen flows through the SARS-CoV-2 recombinant protein (receptor binding domain of spike protein of SARS-CoV-2) labeled with a 40 nm gold (AuNP) particle via chromatographic lateral flow. Then, a complex of IgM-antigen-AuNP/IgG-antigen-AuNP will form. When this complex passes through the coated mouse anti-human IgM/G antibody at M/G coating line, they will show a color band at the coating line (red/purple) forming a double antibody sandwich colloidal gold complex. Here, rabbit IgG conjugated with AuNP will interact with the goat anti-Rabbit IgG antibody, exhibiting a color band at the control line as a standard [19]. Another method is the CRISPR-based method in which a group of Cas nucleases such as cas-12 and cas-13 is used to identify DNA or RNA cleavage sites. This method requires a lateral flow lipstick and can be completed within 40 minutes. The procedure is termed 'SHERLOCK' the target RNA is amplified by Reverse Transcription-Recombinase Polymerase Amplification (RT-RPA) and T7 transcription. The amplified RNA activates cas13. Cas13 then cleaves a reporter RNA cleaving the fluorescent dye from its quencher giving a signal [20]. Cas12 is an RNA-guided DNase that cleaves ssDNA upon binding to its target sequence. In a method called ‘DETECTR’, viral RNA is converted to DNA and then amplified. In the amplified DNA, a specific sequence activates cas12a which subsequently cleaves a reporter molecule containing a fluorophore and quencher molecule. These methods are quite time-saving rather than RT- PCR. Another method is Reverse Transcriptase-Loop-Mediated Isothermal Amplification (RT-LAMP). In this method, DNA polymerase and four to six primers bind to six distinct regions. In a four-primer system, two inner and two outer primers are used. The two internal primers are forward and reverse primers. In a LAMP test sample from a patient is added to a tube and DNA is amplified. This detection depends on turbidity (formation of a byproduct), color (addition of a pH-sensitive dye), or fluorescence (addition of a double-strand binding fluorescent dye). LAMP is a highly sensitive method and reaction occurs in <1 h at 60°C-65°C. This test usually detects 75 copies of RNA per μl [21]. As there is a shortage of kits and false-negative results appear on RT-PCR, chest CT scans are performed. This process is non-invasive and cross- sectional images are produced by taking X-rays at different angles. A radiologist usually analyzes the result and looks for abnormal features. These image features are diverse and depend on the stage of infection after the onset of symptoms in the case of COVID-19 [21]. The sensitivity of chest CT scans is greater than RT-PCR because of the immature development of nucleic acid detection technology, variation in detection, low viral titer, and improper clinical sampling [22]. Among all these methods for COVID-19, detection of viral RNA is convenient with CRISPR-based test and easier than RT-PCR as no bulky instruments are needed. IgM/IgG point of care lateral flow testing is the most straightforward and most rapid method. But professional skill is needed [19].
Around 180 vaccine candidates have been designed so far against SARS-CoV-2 and 71 vaccine candidates are in a clinical trial in humans. Eight vaccines have been authorized for early or limited use and four vaccine candidates (Pfizer-Biontech, Oxford- Astrazeneca, Sputnik-V, and Moderna) have already been approved for full use. Twenty vaccine candidates are in phase III trial with large-scale efficacy test. Twenty-seven vaccine candidates are phase II trials are including their expanded safety trial. Forty vaccine candidates are now testing their safety and dosages are in phase I trial currently. Among all these four vaccine candidates have been abandoned so far because of their inability to trigger an immune response compared to the natural infection or with low immune response and showing unwanted signs and symptoms including testing positive for HIV.
The inactivated vaccine is produced mainly by growing the SARS-CoV-2 virus in Vero cell culture followed by chemical inactivation whereas live attenuated vaccine is based on a genetically weakened virus of SARS-CoV-2, growing the virus in an unfavorable condition (Growth at low temperature) or by rational modification (deleting genes that encounter immune system or by codon de-optimization) (Figure 1). The inactivated vaccine is injected with an adjuvant called “alum (Aluminum hydroxide)”, which generally boosts the immune system to develop antibodies not only against the spike protein but also against the matrix, envelope protein, and nucleocapsid. However, the production of the virus is limited and requires a biosafety level 3 laboratories. Examples include Coronavac, currently under development by Sinovac Biotech, China [11,23]. The live-attenuated vaccine is administered intranasally as it’s the natural route of entry for the virus and provokes the mucosal immune system. Though the virus grows to a limited extent but induces similar responses as caused by the natural infection and antibodies target both structural and non-structural proteins of the virus [24,25]. Among three codon de- optimized vaccines, one is developed by Codagenix in collaboration with the Serum Institute of India [11].
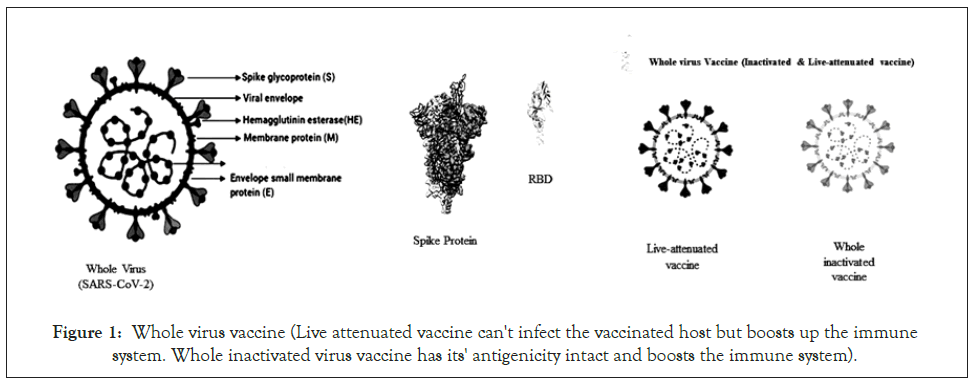
Figure 1: Whole virus vaccine (Live attenuated vaccine can't infect the vaccinated host but boosts up the immune system. Whole inactivated virus vaccine has its' antigenicity intact and boosts the immune system).
This vaccine is engineered to be expressed in different types of expression system without handling the live virus and includes recombinant spike protein-based (deletion of the polybasic cleavage site, the inclusion of two or more stabilizing mutations, the inclusion of trimerizations domains) RBD-based, and virus- like particle-based vaccines (Figure 2) [24,26,27]. The expression system includes eukaryotic expression systems like yeast cells, insect cells, mammalian cells, and plant cells whereas for RBD, the prokaryotic E. coli cells have also been used [11]. The development of spike protein-based and RBD-based vaccines is currently done by Novavax [11].
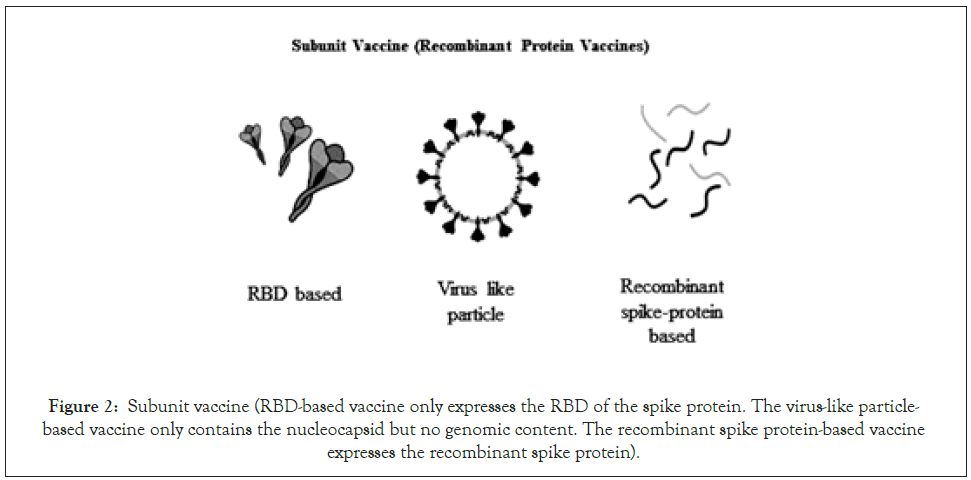
Figure 2: Subunit vaccine (RBD-based vaccine only expresses the RBD of the spike protein. The virus-like particle- based vaccine only contains the nucleocapsid but no genomic content. The recombinant spike protein-based vaccine expresses the recombinant spike protein).
Several virus vectors like adenovirus, modified Ankara virus, human parainfluenza virus, Sendai viruses are used to produce replication- incompetent vector vaccine in which the viruses expresses the spike protein of SARS-CoV-2, and their in-vivo replication is inhibited by deletion of some genes (Figure 3) [28,29]. This vaccine is generally administered intramuscularly and upon entry encodes the spike protein of SARS-CoV-2. However, due to replication incompetence, this vaccine may cause a lower immune response. The ChAdOx-1 nCoV also known as AZ1222 (based on Chimpanzee adenovirus) is one of the vaccine candidates currently in the trial [11]. However, viral vectors able to replicate in the human body expressing a transgene, mainly spike protein are also under development [30,31].
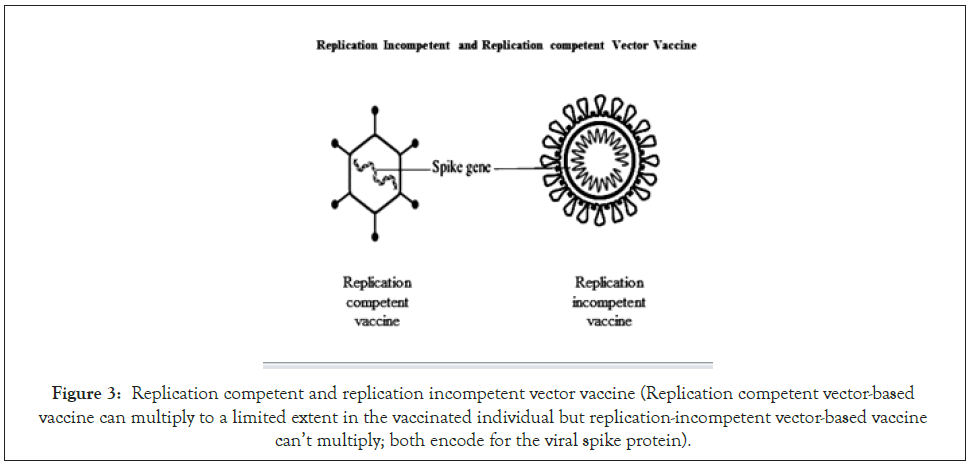
Figure 3: Replication competent and replication incompetent vector vaccine (Replication competent vector-based vaccine can multiply to a limited extent in the vaccinated individual but replication-incompetent vector-based vaccine can’t multiply; both encode for the viral spike protein).
The DNA vaccine generally refers to an engineered spike protein (six variations were used), encoded in a plasmid using either the mammalian expression promoter or virus including a leader sequence i.e. cytomegalovirus promoter to encode the engineered protein that may result in an enhanced immune response (Figure 4) [32,33]. The RNA-based vaccine is a very recent approach. The modified RNA or the self-replicating RNA containing the genetic information for the antigen is delivered with lipid nanoparticles [34]. Pfizer and Moderna have designed these RNA vaccines, which have already got permission for vaccination [35-37].
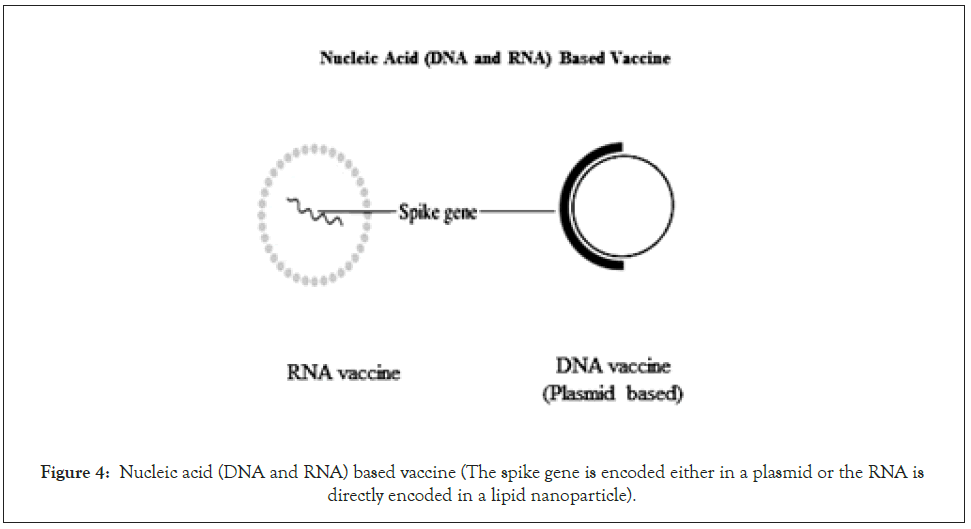
Figure 4: Nucleic acid (DNA and RNA) based vaccine (The spike gene is encoded either in a plasmid or the RNA is directly encoded in a lipid nanoparticle).
The FDA-approved vaccines to date are mostly for the English variant (D614G mutation) and are working effectively against this strain though some suggest that this mutation makes the vaccine less effective. However, the emerging new strain has N501Y mutation that shares an integral part of the spike protein which may cause the vaccination less effective. The UK variant of B.1.1.7 lineage has emerged as a considerable concern because it refracted monoclonal antibodies against the N-terminal and the RBD of the spike protein including convalescent sera vaccine sera [9]. The South African variant of B.1.351 lineage has added more concern because it showed decreased binding with monoclonal antibodies against the Receptor-Binding Motif of the RBD along with the other characteristics [38]. An extensive study is still going on and more research is needed to conclude a final result on the other variants [39]. The vaccine problem may be overcome by using a booster dose tailored to different variants [40].
The world is now facing the most alarming pandemic of this century involving SARS-CoV-2 that has caused immense death. Unremitting mutation if SARS-CoV-2 has once again put the world in a dire situation. Several important mutations caused the virus to transmit faster and reduced binding affinity with the neutralizing antibody, challenging some of the detecting methods. This current study describes the overall mutations of SARS-CoV-2 in different geographical regions, their transmissibility. This study also describes current vaccination strategies along with alternative detection methods that can be useful in detecting different strains of SARS-CoV-This data could help identify differently distributed variants after sequencing and detecting them in a precise manner. The overall study will also aid in understanding the mutations as well as their different clades and lineages.
Citation: Chakrovarty T, Setu AA, Dip SD, Hasan MS, Islam TI (2021) The Emerging Strains of SARS-CoV-2 and Current Vaccine Strategies: A Systemic Review. J Antivir Antiretrovir. S20:001.
Received: 24-Mar-2021 Accepted: 07-Apr-2021 Published: 14-Apr-2021 , DOI: 10.35248/1948-5964.21.s20.001
Copyright: © 2021 Chakrovarty T, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.