
Journal of Clinical and Cellular Immunology
Open Access
ISSN: 2155-9899

ISSN: 2155-9899
Research Article - (2025)Volume 16, Issue 2
Objective: We sought to explore the causal associations between immune cells and Colorectal Cancer (CRC) through the application of Mendelian Randomization (MR).
Methods: The statistical data were gathered from published Genome-Wide Association Studies (GWAS) on immune cells, with genetic variation points being utilized as Instrumental Variables (IVs). For the two-sample Mendelian Randomization (MR) analysis, both Inverse Variance Weighted (IVW) and Bayesian Weighted Mendelian Randomization (BWMR) methods were employed. Additionally, sensitivity analyses were conducted to evaluate the heterogeneity, horizontal pleiotropy and robustness of the findings.
Results: The IVW analysis revealed that 37 immunophenotypes have a causal link to colorectal cancer. Additionally, the BWMR method identified 4 immune phenotypes with a causal connection to the disease’s development. In contrast, Reverse MR analysis demonstrated a significant causal relationship between colorectal cancer and 2 specific immune cells.
Conclusion: This research uncovers, for the first time, the causal link between distinct immune cell traits and the onset of colorectal cancer. It offers a fresh viewpoint for comprehending the immunological underpinnings of the disease. The insights gained are pivotal in devising precise preventive and therapeutic approaches and they bear the potential to enhance the clinical management and treatment success rates for individuals afflicted with colorectal cancer.
Colorectal cancer; Immune cell dynamics; Mendelian Randomization (MR)
Colorectal cancer ranks as the third most prevalent form of cancer and stands as the second leading cause of cancer-related deaths globally, imposing substantial socioeconomic challenges [1-3]. Growing evidence indicates that the Tumor Microenvironment (TME) encompassing tumor cells and their associated immune and stromal cells-exerts a direct influence on tumor biology and the response to immunotherapy [4,5]. Furthermore, the infiltration of immune cells within tumors has been shown to impact the effectiveness of chemotherapy, radiotherapy or immunotherapy, as well as tumor angiogenesis and metastasis. Consequently, these immune cells are being investigated as a crucial target for managing tumor progression [6,7].
Mendelian Randomization (MR) is an analytical technique predominantly used to determine causality. It utilizes Single Nucleotide Polymorphisms (SNPs), which have strong associations with specific phenotypes, as Instrumental Variables (IVs) [8,9]. This method addresses common challenges in observational studies, such as confounding factors and reverse causation [10]. Furthermore, there’s a growing body of evidence highlighting the utility of the human genetic interface in analyzing immune cell phenotypes for clinical research [11-13]. Recent studies underscore this point. For instance, one investigation examined a potential causal link between Autoimmune Diseases (AIDs) and Colorectal Cancer (CRC) [14]. Another study identified the causal influence of immune cells on gastrointestinal cancers [15]. These research efforts employed MR analysis to explore the connections between immune signatures and cancer, complemented by sensitivity analyses to verify the robustness of their findings. In light of this, our objective is to apply MR to deduce causal relationships between 731 immune cell signatures and colorectal cancer, potentially paving the way for new insights in cancer immunology.
Data sources for exposure data
In a comprehensive study involving 3,757 Sardinians, genotyping and imputation were conducted on roughly 22 million SNPs. This was achieved using high-density arrays and a reference panel derived from Sardinian sequences. The GWAS summary statistics for various immune traits are accessible in the GWAS Catalog, with accession numbers ranging from GCST0001391 to GCST0002121. The cataloged immune signatures encompass: 118 phenotypes for Absolute Cell count (AC), 389 phenotypes for Median Fluorescence Intensity (MFI), 32 phenotypes for Morphological Parameters (MP) and 192 phenotypes for Relative Cell count (RC) [11,16].
Data sources for outcome data
The comprehensive dataset on colorectal cancer, which includes 6,509 cases and 287,137 controls, was sourced from the FinnGen database(https://risteys.finregistry.fi/endpoints/ C3_COLORECTAL_EXALcolorectal cancer). The data, corresponding to release 11, was made available on November 2, 2023. FinnGen represents a significant public-private partnership that aggregates and scrutinizes genetic and health-related data from approximately 500,000 participants enrolled in the Finnish Biobank. The database was last accessed on August 8, 2022 [17].
Mendelian randomization design and instrumental variables selection
In our research, Single Nucleotide Polymorphisms (SNPs) were utilized as Instrumental Variables (IVs). To qualify as an IV, a genetic variant must meet the following criteria: (1) The SNP must have a strong association with the exposure; (2) The SNP should not be linked to any confounders; (3) The exposure must be the only mediator of the SNP’s effect on the outcome [18]. Given the scarcity of SNPs that meet the GWAS significance threshold, we adopted a more permissive criterion of P<1 × 10-5 [19]. The selected SNPs also conformed to Linkage Disequilibrium (LD) requirements, with r2<0.001 within a 10,000 kb region and SNPs with palindromic structures were excluded [20]. To assess the strength of the SNPs, we calculated F statistics (F=beta2/se2) for each one. An F-statistic greater than 10 is deemed sufficient to counteract the effects of weak IVs (Fstatistics ≤ 10).
MR analyses
Our study utilized a variety of statistical methods, including: Inverse Variance Weighted (IVW), MR-Egger, weighted median, weighted mode, simple mode and MR-Pleiotropy Residual Sum and Outlier (MR-PRESSO) tests. These approaches were critical in addressing the challenges posed by the polygenic nature of complex immune traits and the frequent presence of pleiotropy. To navigate these complexities, we further employed the Bayesian Weighted Mendelian Randomization (BWMR) method for causal inference. This technique leverages genetic variants as Instrumental Variables (IVs) to estimate the causal impact of one phenotype on another. Furthermore, our research incorporated reverse MR analysis to explore the potential causal effects of Colorectal Cancer (CRC) on immune cell phenotypes.
Sensitivity analysis
Heterogeneity in our analysis was evaluated using Cochran’s Q test, with a p-value less than p<0.05 indicating significant heterogeneity. The presence of pleiotropic effects was examined through the MR-Egger intercept test. An intercept close to zero or a p-value greater than p>0.05 was interpreted as an absence of pleiotropy in the Instrumental Variables (IVs). To mitigate the risk of horizontal pleiotropy stemming from any single SNP, we conducted leave-one-out analysis.
Statistical analysis
To assess the causal relationship between the variables, we employed the Odds Ratio (OR) along with its 95% Confidence Interval (CI). Our analysis adhered to established Mendelian Randomization (MR) guidelines. The computational work was carried out in R, utilizing packages such as TwoSampleMR, ggplot2, MVMR and MR-PRESSO for the analysis.
Ethics statement
The data employed in our analysis are publicly accessible and have received approval from the institutional review committees of the respective studies. Consequently, no additional ethical approvals were required. The outcomes of our analysis are comprehensively detailed in the main article and its supplementary materials.
The causal role of immunophenotypes on colorectal cancer
We first investigated the interrelationships between 731 immune cell signatures and colorectal cancer with the IVW method being the primary analytical approach employed. With a significance level of 0.05, we detected 37 indicative immunophenotypes. The analysis is shown in Figure 1. Further analysis using the Bayesian Weighted Mendelian Randomization (BWMR) method, due to its restrictive nature, pinpointed 4 significant immunophenotypes, including CD25hi CD45RA+ CD4 not Treg AC (95% CI: 0.887-0.971, P=0.001), Basophil CD33dim HLA DR- CD66b- (95% CI: 0.906-0.984, P=0.005), EM DN (CD4-CD8-) %T cell (95% CI: 1.021-1.143, P=0.007) and FSC-A on CD14+ monocyte (95% CI: 0.893-0.987, P=0.013). The detailed results are presented in Table 1. Mendelian randomization analyses of the causal effects between immunophenotypes and colorectal cancer by IVW methods.
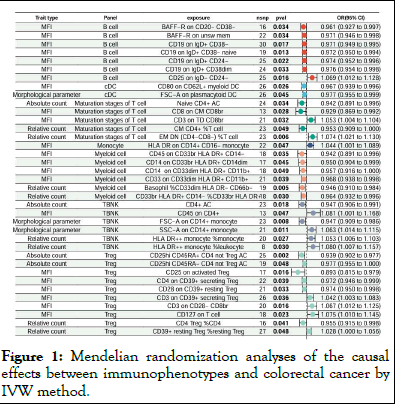
Figure 1: Mendelian randomization analyses of the causal effects between immunophenotypes and colorectal cancer by IVW method.
Panel |
Trait |
Method |
or |
or_lci95 |
or_uci95 |
pval |
msT cell |
EM DN (CD4-CD8-) %T cell |
BWMR |
1.081 |
1.021 |
1.144 |
0.007 |
Myeloid cell |
Basophil %CD33dim HLA DR- CD66b- |
BWMR |
0.944 |
0.906 |
0.984 |
0.006 |
TBNK |
FSC-A on CD14+ monocyte |
BWMR |
0.939 |
0.893 |
0.987 |
0.014 |
Treg |
CD25hi CD45RA+ CD4 not Treg AC |
BWMR |
0.928 |
0.887 |
0.971 |
0.001 |
Table 1: Mendelian randomization analyses of the causal effects between immunophenotypes and colorectal cancer by MWMR method.
or, odds ratio; or_lci95, lower limits of 95% confidence intervals; or_uci95, upper limits of 95% confidence interval. BWMR, Bayesian Weighted Mendelian Randomization; msT cell, maturation stages of T cell.
The causal effect of colorectal cancer on immunophenotypes
In the reverse MR analysis, IVW methodology indicated causal links between colorectal cancer and specific immunophenotypes: CD25 on IgD- CD24- (95% CI: 0.773-0.974, P=0.016) and CD14 on CD33br HLA DR+ CD14dim (95% CI: 0.686-0.965, P=0.017); The BWMR approach corroborated these findings: CD25 on IgD- CD24- (95% CI: 0.771-0.977, P=0.018) and CD14 on CD33br HLA DR+ CD14dim (95% CI: 0.684-0.967, P=0.019). These results are detailed in Table 2.
Panel |
Trait |
Method |
or |
or_lci95 |
or_uci95 |
pval |
B cell |
CD25 on IgD- CD24- |
IVW |
0.868 |
0.773 |
0.974 |
0.016 |
B cell |
CD25 on IgD- CD24- |
BWMR |
0.868 |
0.771 |
0.977 |
0.019 |
Myeloid cell |
CD14 on CD33br HLA DR+ CD14dim |
IVW |
0.813 |
0.686 |
0.965 |
0.018 |
Myeloid cell |
CD14 on CD33br HLA DR+ CD14dim |
BWMR |
0.813 |
0.684 |
0.967 |
0.019 |
Table 2: Mendelian randomization analyses of the causal effects of colorectal cancer on immunophenotypes by IVW and MWMR methods.
or, odds ratio; or_lci95, lower limits of 95% confidence interval; or_uci95, upper limits of 95% confidence intervals. BWMR, Bayesian Weighted Mendelian Randomization; IVW, inverse variance weighted.
Sensitivity analysis
The robustness of the above results was confirmed as no horizontal pleiotropy was detected. Cochran’s Q test showed no heterogeneity across all outcomes. The leave-one-out analysis further validated the unbiased nature of our MR estimates. The consistency of our findings is illustrated in the scatter and funnel plots.
Based on a large amount of publicly available genetic data, we explored the causal relationship between 731 immune cell features and colorectal cancer. To our knowledge, this is the first Mendelian randomized analysis of the causal relationship between multiple immune cell phenotypes and colorectal cancer. Among the seven types of the immune panel (B cell, cDC, maturation stages of T cell, monocyte, myeloid cell, TBNK and Treg), 37 immune cell phenotypes were found to have significant causal effects on colorectal cancer and colorectal cancer led to changes in 2 immunophenotypes.
Tumor Immune Microenvironment (TIME) is closely related to tumor progression. CD8+ T cells destroy tumor cells by binding to Major Histocompatibility Complex (MHC) class I-restricted epitopes, which is positively correlated with more favorable clinical outcome of colorectal cancer. A recent study of most tumor-infiltrating immune cell subtypes revealed that CD8+ T cells had the greatest impact on patient survival. Meanwhile, CD4+ T cells can eradicate tumors through directly cytolytic activity or indirectly mediating potent cytotoxic anti-tumor effects. T cell absence is believed to be a reason why tumor shrinkage is minimally induced by immune checkpoint inhibitors. Several studies indicate that a high degree of infiltration of cDC (myeloid/conventional DC1 (cDC1), myeloid/conventional DC2 (cDC2) into tumors was possibly able to promote immune response. Our research is in agreement with the notion that CD8+ or CD4+ T cell, CD80 on CD62L+ myeloid DC, and FSC-A on plasmacytoid DC infiltration is associated with a better prognosis.
Interestingly, the infiltration of myeloid cells has been associated with both good and poor outcomes of colorectal cancer. Juha et al., reported that higher densities of both intraepithelial (P trend=0.0002) and stromal (P trend<0.0001) CD14HLA-DR mature monocytic cells were associated with lower cancer-specific mortality while, conversely, higher intraepithelial densities of CD14HLA-DR+++− immature monocytic cells were associated with higher cancer-specific mortality (P trend=0.0003). However, this finding is not consistent with ours.
Tregs represent a heterogeneous cell population in the colorectal cancer microenvironment that can stimulate and suppress immune responses depending on their phenotype expressions. CD4CD25Foxp3 regulatory T cells is believed to facilitate sequestration of IL-2 from the environment, thereby reducing effector T-cell access to IL-2. Research has shown that higher expression of CD39 in Treg cells correlated with a poorer outcome. While our research reported that their presence predicts both good and poor outcomes of colorectal cancer. A likely explanation of these conflicting reports could be the coexistence of phenotypically similar Treg subsets which nonetheless have different functions.
In addition, the study also conducted reverse MR analysis (using colorectal cancer as the exposure and immune cell traits as the outcome) and confirmed that colorectal cancer significantly changed the phenotype and function of immune cells.
Our study has several strengths. Firstly, it is the most systematic study to date on the causal relationship between immune cell signatures and colorectal cancer risk. When involved a lot of data analysis, we also employed the recently developed BWMR method. BWMR can not only account for the uncertainty of estimated weak effects and weak horizontal pleiotropic effects but also adaptively detect outliers due to a few large horizontal pleiotropic effects. To ensure reliable estimates, a series of sensitivity analyses and implemented stringent quality control measures were used to ensure the accuracy of our results.
However, this work also has some limitations. Firstly, because only people of European heritage were included in the GWAS, the conclusions of this study might not apply to people of other racial or ethnic backgrounds. Future studies must incorporate a more diverse population to improve the generalizability of this results. Secondly, we set a more relaxed threshold threshold (p<1e-5) for SNP selection. Meanwhile, we combined with sensitivity analysis and robustness tests to increase the strength of the evidence. Thirdly, our findings necessitate subsequent verification in real-world study and wet lab experimentation. However, the experimental validations have been hindered by cost, resources and cumbersome technical expertise required for cancer immunomics investigations. Further research is also indispensable to explore the associations between the immune repertoire of CRC and the intestinal microbiota, metabolomics, which is essential for the development of novel immune therapies for CRC.
In conclusion, we demonstrated the causal relationship between several immune phenotypes and CRC through MR analysis, highlighting the complex interaction patterns between the immune system and CRC.
This work was made possible by the generous sharing of GWAS summary statistics from the FinnGen consortium and UK Biobank. We thank all the individual patients who provided the sample that made the data available and all the investigators who provided these data to support this study.
The authors have no financial conflicts of interest.
LH and YH conceived and designed the study. YH and LH are responsible for data collection and compilation, data analysis, interpretation and manuscript writing. YH and LH revised the manuscript. All work is performed under LH and YH instructions. All authors contributed to the article and approved the submitted version.
This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
Not applicable.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholars] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Huang L, Huang Y (2025) The Causal Role of Immunophenotypes in Colorectal Cancer: A Mendelian Randomization Study. J Clin Cell Immunol. 16:752.
Received: 11-Apr-2024, Manuscript No. JCCI-24-30742; Editor assigned: 15-Apr-2024, Pre QC No. JCCI-24-30742 (PQ); Reviewed: 29-Apr-2024, QC No. JCCI-24-30742; Revised: 02-Apr-2025, Manuscript No. JCCI-24-30742 (R); Published: 09-Apr-2025 , DOI: 10.35248/2155-9899.25.16.752
Copyright: © 2025 Huang L, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.