Fungal Genomics & Biology
Open Access
ISSN: 2165-8056
ISSN: 2165-8056
Research Article - (2022)Volume 12, Issue 4
The circumstances in which organisms live induce polymorphism in their genes, including fungal allergen genes, leading to altered structures and functions of proteins, related to their pathogenicity. Major allergen genes of Aspergillus fumigatus, Asp f 1, Asp f 2, and Asp f 3, were examined in 59 strains [environment and animal/human- body origin] to determine their nucleotide sequences, and then categorized. The location and number of IgE epitopes on the allergen molecules were predicted using computer software. The Asp f 1 gene was classified into two groups (f1-1 and f1-2). One of the groups possessed one-nucleotide mutation point with one amino-acid substitution. The mutated Asp f 2 gene accompanying 6-amino acid substitution was classified into 7 groups (f2-1 to f2-7). Six of the groups possessed a newborn IgE epitope. The Asp f 3 gene contained two mutations, resulted in three groups (f3-1 to f3-3) without any amino-acid substitutions. Category E, consisting of group’s f1-1, f2-5, and f3-2, was specific to an environmental origin. Our findings suggest that nucleotide mutation of the fungal allergen genes, associated with the origin of the fungus, modifies the structure of proteins, and affects their pathogenic properties, such as the localization of IgE epitopes.
Aspergillus fumigatus; IgE; Allergenic proteins; Allergen genes
Fungi cause human diseases, such as infections, mycotoxicosis, and allergies. It is believed that fungi are ubiquitous airborne allergens. More than 80 fungi genera are recognized to be associated with allergenic symptoms [1]. The genera Alternaria, Cladosporium, Penicillium, Aspergillus, and Malassezia have been well studied [2]. A. fumigatus is ranked the top pathogenic fungus. A. fumigatus causes infections and allergies, such as allergic rhinitis, asthma, sinusitis, and Allergic Bronchopulmonary Aspergillosis (ABPA) [3,4].
Allergenic proteins are defined as proteins that are capable of stimulating the production of Immunoglobulin E (IgE) antibodies in the human body and to specifically bind to IgE antibodies, which exist in the sera of allergic patients. A candidate protein is identified as an allergen after epidemiological, etiological, and immunological examinations. Once the gene of such protein is cloned, and sequenced, it is registered in the International Union of Immunological Societies allergen database (IUIS allergen database; URL http://www.allergen.org/).
A. fumigatus allergenicity is also well analyzed [1,2], and a total of 23 A. fumigatus allergens are listed in the IUIS allergen database. Several of these, namely Asp f 1, Asp f 2, and Asp f 3, are recognized as major A. fumigatus allergens [5-8]. A total of 85% of patients with ABPA or asthma who showed a positive immunoglobulin E (IgE) antibody titer against the extracted allergens of A. fumigatus possessed an IgE antibody against the purified and recombinant Asp f 1 protein [6]. Asp f 2 is an allergic protein with a molecular weight of 37 kDa. Both isolated and recombinant Asp f 2 protein bound to the IgE antibody in sere of ABPA and cystic fibrosis-ABPA patients [7]. Asp f 3 allergen, which was cloned in 1997, is a protein composed of 168 amino acids. The binding ability of IgE antibodies to recombinant Asp f 3 was detected in 72% of 89 patients who were positive for a skin test using the crude allergen extracted from an A. fumigatus culture [8].
Infection causes modifications in the cellular and molecular events occurring between hosts and pathogens. Such interactions may be one of the pressures that lead to many kinds of alterations of organism properties, via nucleotide mutation of genes. In addition, the origin of microorganisms is an important factor in mutations. Environmental pressure induces epigenetic and genetic modifications on all creatures, including fungi.
We are interested in the influence of the origin of the isolated fungi on the genes of allergens, which affect the immunological conditions of humans. The objective of this study was to examine the nucleotide mutations in allergen genes, Asp f 1, Asp f 2, and Asp f 3, of A. fumigatus isolated from the environment, such as house dust, room air, and soil, and from the body of animals/humans, resulting from the colonization of A. fumigatus after infection. We discuss the probability that the origin-specific adaption of allergen genes makes fungus more allergenic.
Fungal strains
A total of 59 strains of A. fumigatus, including 29 strains from different environments [from room air (2), house dust (12), and soil (15)], 13 from infectious lesions [from humans (6) and animals (7)] and 17 of unknown source, were used in this study. The strains were collected in several areas in Japan, and sent to, and maintained in the mycological laboratory of the National Institute of Health Sciences (Kawasaki, Japan).
Identification of the fungal isolates
All stocked isolates were inoculated on malt extract agar (Oxoid, Thermo Scientific, Wilmington, DE) and Czapek agar containing 3.5% Czapek-dox broth (Becton, Dickinson and Company, Franklin Lakes, NJ) and 1.5% agar, and incubated at 25°C for 10 days. The Aspergillus strains were examined and identified using the general method by Raper et al., with a focus on morphology [9]. All strains were cultured on a potato dextrose agar (Eiken, Tokyo, Japan) slant medium at 25°C for 10 days. Mycelia or conidia were harvested from the slant culture in a tube containing 1 ml of potato dextrose broth (Becton, Dickinson and Company), and incubated at 25°C for 3 days. These cultures were centrifuged at 18,000 × g for 10 minutes. Genomic DNA was extracted from the pellets using the sodium dodecyl sulfate (SDS) method with minor modifications [10]. Briefly, an 1-ml aliquot of a lysing buffer containing 50 mM Tris-HCl buffer (pH 8.0), 250 mM NaCl, 50 mM EDTA, 0.3% SDS, and 20 μl of RNase A (10 mg/ml; Novagen, Darmstadt, Germany) were added to the tube containing the fungal body pellets, and mixed vigorously. The upper water-phase containing genomic DNA was harvested. The genomic DNA was precipitated using chilled isopropanol, and suspended in 20 μl of Tris-EDTA buffer, (pH 7.5). The extracted genomic DNA was measured spectrophotometrically to examine its quality and concentration using a NanoDrop 1000 Spectrophotometer V3.7 (Thermo Fisher Scientific, Wilmington, DE).
The β-tubulin gene was used in this study as the identification marker gene of A. fumigatus. A fragment of the β-tubulin gene was amplified using the primer pair Bt2a (5’-GGTAACCAAATCGGTGCTGCTTTC-3’) and Bt2b (5’-ACCCTCAGTGTAGTGACCCTTGGC-3’) [11]. TaKaRa ExTaq (TaKaRa Bio Inc., Otsu, Japan) was used according to the manufacturer’s instructions for amplification in a thermal cycler (GeneAmp PCR System 9700; Applied Biosystems, Foster City, CA). The PCR program consisted of an initial denaturing step at 94°C for 5 minutes, 35 amplification cycles (94°C for 30 seconds, 60°C for 40 seconds, and 72°C for 1 minute), and an additional extension step at 72°C for 3 minutes. The PCR products were purified using ExoSap-IT (USB; Cleveland, OH), and then directly sequenced. The obtained two sequences were concatenated into one sequence using a sequence assembly software program, ATGC (Genetyx Corporation, Tokyo, Japan). The determined sequences were entered into the Basic Local Alignment Search Tool (BLAST) for comparison with the sequences registered in the nucleotide database of the National Center for Biotechnology Information. When the result of a sequence homology search of the isolates indicated a high homology with the A. fumigatus β-tubulin gene, the isolate was identified as A. fumigatus.
The determination of the nucleotide sequence of Aspergillus fumigatus allergen gene
The fragments of these genes that contained a coding region sequence were amplified and sequenced using the following primer pairs, which were designed using a publicly available software program, Primer 3 Plus (http://www.bioinformatics.nl/cgi-bin/ primer3plus/primer3plus.cgi/), based on the sequences of Accession Nos. M83781 (Asp f 1), U56938 (Asp f 2), and U58050 (Asp f 3).
The primer sequences used for the Asp f 1 gene were as follows-
Aspf1-F: 5’-TGCCCTGACTACGTCCAAG-3’
Aspf1-R: 5’-CCTCCTAAGGCATAATACCCACA-3’
The primer sequences used for the Asp f 2 genes were as follows
Aspf2-F: 5’-TCACGCACCATCCAACCT-3’
Aspf2-R: 5’-GCGTTAACATACGGCGTCAA-3’
The primer sequences used for the Asp f 3 genes were as follows-
Aspf3-F: 5’-ATGTCTGGACTCAAGGCCGGTGACAG-3’
Aspf3-R: 5’-TCCCATGCCGGCATCTATCATGTCTAGTT-3’
The conditions used for the amplification were the same as those used for the β-tubulin gene (described above), with slight modifications. The PCR program consisted of an initial denaturing step at 94°C for 5 minutes, 35 amplification cycles (94°C for 30 seconds, 60°C for 40 seconds, and 72°C for 40 seconds), and an additional extension step at 72°C for 3 minutes. The amplicons were directly sequenced.
Prediction of IgE epitopes and the three-dimensional structure models of Asp f 1, Asp f 2, and Asp f 3 proteins
The location of IgE epitopes on Asp f 1, Asp f 2, and Asp f 3 was predicted using the Linear Epitope Prediction Based on the Propensity Scale ASM software program (http://leps.cs.ntou. edu.tw/). The query for the software was the deduced amino acid sequence of the three allergens that were examined in this study.
Polymorphic properties of Aspergillus fumigatus allergen genes, Asp f 1, Asp f 2, and Asp f 3
A total of 57 strains (96.6%) of the 59 isolates showed the sequence of Asp f 1 gene, same as that registered in the IUIS database. The other two strains of the Asp f l gene showed one nucleotide mutation at the 262 bp position (Table 1). This mutation resulted in an amino acid substitution at the 88th residue. These data indicate that the Asp f 1 gene is classified into two groups: f1-1 and f1-2 (Table 1). The sequencing of the tested strains of the Asp f 2 genes indicated 10 mutation points accompanying amino acid substitutions (Table 1). The nucleotide sequence of Asp f 2 genes in the IUIS database was not found among any of the 59 strains tested. The Asp f 2 gene of the 59 strains was classified into seven groups, based on the combination of mutation points (Table 1). A total of 42 of the 59 strains (71.2%) belonged to the f2-2 group. This dominant group contained four mutation points, which were associated with three amino acid substitutions (Table 1). The f2-5 group (12 strains, 20.0%) was the next dominant group. It contained one more mutation point than the f2-2 group. The Asp f 3 genes of the 59 strains showed three distinct nucleotide sequences. A total of 18 (30.5%) of the 59 strains corresponded to the nucleotide sequence of the Asp f 3 gene registered in the IUIS database (group f3-1). Of the 59 strains, 34 (57.6%) showed one mutation in the sequence at 147 bp (group f3- 2). This mutation group was dominant. Group f3-3 possessed one more mutation point than group f3-2, which was a mutation at 240 bp (Table 1). Neither of these two nucleotide mutations affected the amino acid sequence.
| Allergen name | Group name | Number of strains | Number of point mutations | Position (bp) and aspect of single nucleptide substitution | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Position and aspect of amino acid substitution | ||||||||||||||
| Asp f 1 | f1-1 | 57 | 0 | ー 2) | ||||||||||
| ー | ||||||||||||||
| f1-2 | 2 | 1 | 262bp (C→T) | |||||||||||
| L88F | ||||||||||||||
| Asp f 2 | f2-1 | 1 | 3 | 681bp (C→G) | 682bp (G→C) | 720bp (T→C) | ||||||||
| F227L | E228Q | ー | ||||||||||||
| f2-2 | 42 | 4 | 681bp (C→G) | 682bp (G→C) | 720bp (T→C) | 889bp (T→C) | ||||||||
| F227L | E228Q | ー | S297P | |||||||||||
| f2-3 | 1 | 5 | 402bp (C→T) | 681bp (C→G) | 682bp (G→C) | 720bp (T→C) | 889bp (T→C) | |||||||
| ー | F227L | E228Q | ー | S297P | ||||||||||
| f2-4 | 1 | 5 | 476bp (G→A) | 681bp (C→G) | 682bp (G→C) | 720bp (T→C) | 889bp (T→C) | |||||||
| R159H | F227L | E228Q | ー | S297P | ||||||||||
| f2-5 | 12 | 5 | 681bp (C→G) | 682bp (G→C) | 720bp (T→C) | 827bp (G→C) | 889bp (T→C) | |||||||
| F227L | E228Q | ー | G276A | S297P | ||||||||||
| f2-6 | 1 | 6 | 85bp (G→A) | 426bp (G→A) | 681bp (C→G) | 682bp (G→C) | 720bp (T→C) | 889bp (T→C) | ||||||
| A29T | ー | F227L | E228Q | ー | S297P | |||||||||
| f2-7 | 1 | 6 | 681bp (C→G) | 682bp (G→C) | 720bp (T→C) | 825bp (G→A) | 827bp (G→C) | 889bp (T→C) | ||||||
| F227L | E228Q | ー | ー | G276A | S297P | |||||||||
| Asp f 3 | f3-1 | 18 | 0 | ー | ||||||||||
| ー | ||||||||||||||
| f3-2 | 34 | 1 | 147bp (T→C) | |||||||||||
| ー | ||||||||||||||
| f3-3 | 7 | 2 | 147bp (T→C) | 240bp (T→C) | ||||||||||
| ー | ー | |||||||||||||
Note: 1) DNA sequence of the allergen of each test strain was compared with the reference sequence opened in IUIS allergen nomenclature (http://www.allergen.org/index.php). 2) No substitution
Table 1: Mutation of the nucleotides and alteration of the amino acid sequence of Aspergillus fumigatus allergens and grouping of the genes based on the mutations1).
Categorization of the groups of the allergen genes of Aspergillus fumigatus and their relationship to their origin
Categories A to G were recognized based on a combination of the groups of Asp f 1, Asp f 2, and Asp f 3 in the total of 42 strains whose origin was clarified (Table 2). Categories C, D, F, and G were minor because of the markedly small number of strains, suggesting the importance of Categories A, B, and E. Categories A and B consisted of the isolates whose origin was environment or disease (animal/ human body). However, category E consisted of isolates derived from the environment (Table 2). All isolates of A. fumigatus were classified, according to the category and group in Table 3. Group f3-2 existed in categories B and E. Group f3-2 was related to group f2-2 in category B, and with f2-5 in category E. Group f2-2 in category B consisted of the isolates from disease and environment origins, but group f2-5 specifically consisted of the isolates from the environment origin.
| Category | Group name | Number of strain | ||||
|---|---|---|---|---|---|---|
| Asp f 1 | Asp f 2 | Asp f 3 | Total (%) | Environment | Disease | |
| A | f1-1 | f2-2 | f3-1 | 11 (26) | 7 | 4 |
| B | f1-1 | f2-2 | f3-2 | 16 (38) | 10 | 6 |
| C | f1-1 | f2-2 | f3-3 | 2 (5) | 0 | 2 |
| D | f1-1 | f2-3 | f3-1 | 1 (2) | 1 | 0 |
| E | f1-1 | f2-5 | f3-2 | 10 (24) | 10 | 0 |
| F | f1-1 | f2-6 | f3-3 | 1 (2) | 0 | 1 |
| G | f1-1 | f2-7 | f3-2 | 1 (2) | 1 | 0 |
Table 2: Categolyzation of Aspergillus fumigatus based on their grouping and number of the tested isolates in the categolies, depending on their origins.
| Category | Origin | Group of allergen genes | ||
|---|---|---|---|---|
| Asp f 1 | Asp f 2 | Asp f 3 | ||
| A | Disease_animal | f1-1 | f2-2 | f3-1 |
| Disease_animal | f1-1 | f2-2 | f3-1 | |
| Disease_human | f1-1 | f2-2 | f3-1 | |
| Disease_human | f1-1 | f2-2 | f3-1 | |
| B | Disease_animal | f1-1 | f2-2 | f3-2 |
| Disease_animal | f1-1 | f2-2 | f3-2 | |
| Disease_animal | f1-1 | f2-2 | f3-2 | |
| Disease_animal | f1-1 | f2-2 | f3-2 | |
| Disease_human | f1-1 | f2-2 | f3-2 | |
| Disease_human | f1-1 | f2-2 | f3-2 | |
| C | Disease_human | f1-1 | f2-2 | f3-3 |
| Disease_human | f1-1 | f2-2 | f3-3 | |
| F | Disease_animal | f1-1 | f2-6 | f3-3 |
| A | Environment_house dust | f1-1 | f2-2 | f3-1 |
| Environment_house dust | f1-1 | f2-2 | f3-1 | |
| Environment_house dust | f1-1 | f2-2 | f3-1 | |
| Environment_house dust | f1-1 | f2-2 | f3-1 | |
| Environment_soil | f1-1 | f2-2 | f3-1 | |
| Environment_soil | f1-1 | f2-2 | f3-1 | |
| Environment_soil | f1-1 | f2-2 | f3-1 | |
| B | Environment_room air | f1-1 | f2-2 | f3-2 |
| Environment_house dust | f1-1 | f2-2 | f3-2 | |
| Environment_house dust | f1-1 | f2-2 | f3-2 | |
| Environment_soil | f1-1 | f2-2 | f3-2 | |
| Environment_soil | f1-1 | f2-2 | f3-2 | |
| Environment_soil | f1-1 | f2-2 | f3-2 | |
| Environment_soil | f1-1 | f2-2 | f3-2 | |
| Environment_soil | f1-1 | f2-2 | f3-2 | |
| Environment_soil | f1-1 | f2-2 | f3-2 | |
| Environment_soil | f1-1 | f2-2 | f3-2 | |
| D | Environment_room air | f1-1 | f2-3 | f3-1 |
| E | Environment_house dust | f1-1 | f2-5 | f3-2 |
| environment_house dust | f1-1 | f2-5 | f3-2 | |
| Environment_house dust | f1-1 | f2-5 | f3-2 | |
| Environment_house dust | f1-1 | f2-5 | f3-2 | |
| Environment_house dust | f1-1 | f2-5 | f3-2 | |
| Environment_house dust | f1-1 | f2-5 | f3-2 | |
| Environment_soil | f1-1 | f2-5 | f3-2 | |
| Environment_soil | f1-1 | f2-5 | f3-2 | |
| Environment_soil | f1-1 | f2-5 | f3-2 | |
| Environment_soil | f1-1 | f2-5 | f3-2 | |
| G | Environment_soil | f1-1 | f2-7 | f3-2 |
Table 3: Distribution of the group of Asp f 1, Asp f 2, and Asp f 3 allergen genes among the individulal isolates of Aspergillus fumigatus.
The influence of amino-acid substitution of Aspergillus fumigatus allergen proteins on their IgE epitopes
The properties of Asp f 1 protein, regarding the amino-acid substitution resulting from polymorphisms of the nucleotide sequence and the information for its IgE epitopes are summarized in Figure 1A. The dominant f1-1 group (57 of 59 strains), which possessed a sequence that was the same as that registered in the IUIS database, contained five IgE epitope sites (Figure 1B) on the Asp f 1 molecule. The two stains (3.4%) that belonged to the f1-2 group included a mutated nucleotide sequence, resulting in an amino acid substitution at the 88th position and a deletion of the IgE epitope (Figures 1B and 1C). The three-dimensional structure is shown in Figure 1D.
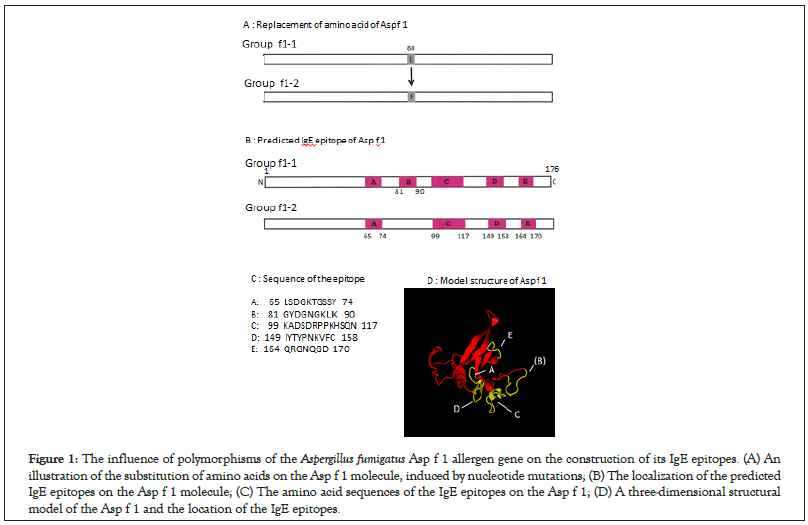
Figure 1: The influence of polymorphisms of the Aspergillus fumigatus Asp f 1 allergen gene on the construction of its IgE epitopes. (A) An illustration of the substitution of amino acids on the Asp f 1 molecule, induced by nucleotide mutations; (B) The localization of the predicted IgE epitopes on the Asp f 1 molecule; (C) The amino acid sequences of the IgE epitopes on the Asp f 1; (D) A three-dimensional structural model of the Asp f 1 and the location of the IgE epitopes.
The properties of the Asp f 2 allergen, regarding the amino acid substitution and the information for its IgE epitope sites are summarized in Figure 2A. Although the nucleotide mutation occurred at 10 positions of the Asp f 2 genes, four of these mutations did not influence the amino acid sequence. The other 6 mutations, however, replaced the amino acids. One strain of the f2-1 group, in which there was no replacement at position 297th, revealed four IgE epitopes (Figure 2A, epitopes A to D). Group f2-2, which was dominant (42 of 59 stains), contained five IgE epitope sites (Figure 2A, epitopes A to E). Although the amino acid sequences of the f2-3, f2-4, f2-5, f2-6, and f2-7 groups were different from that of the dominant f2-2 group, the construction properties of the IgE epitopes were the same as those of group f2- 2. A new epitope (E) was induced in the Asp f 2 protein of group f2-5 (Figures 2A-2D).
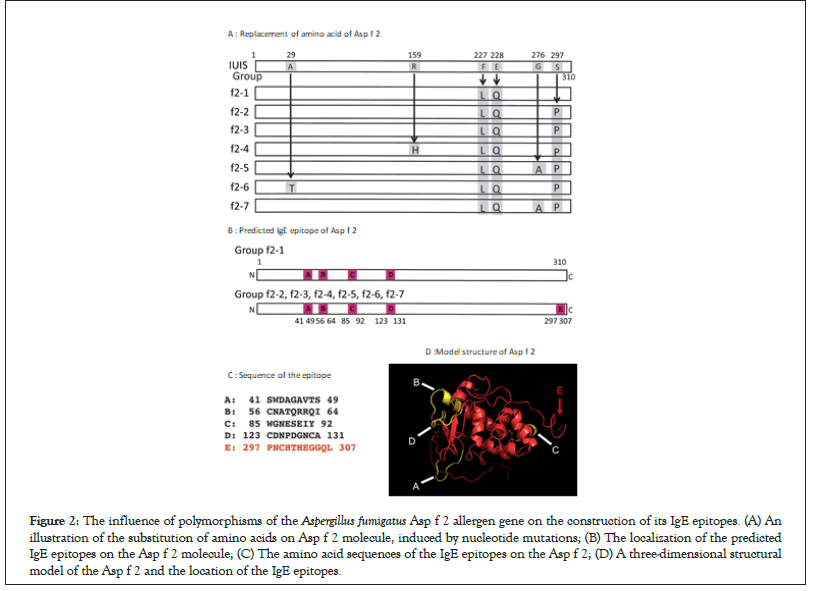
Figure 2: The influence of polymorphisms of the Aspergillus fumigatus Asp f 2 allergen gene on the construction of its IgE epitopes. (A) An illustration of the substitution of amino acids on Asp f 2 molecule, induced by nucleotide mutations; (B) The localization of the predicted IgE epitopes on the Asp f 2 molecule; (C) The amino acid sequences of the IgE epitopes on the Asp f 2; (D) A three-dimensional structural model of the Asp f 2 and the location of the IgE epitopes.
The properties of the Asp f 3 allergen proteins, regarding the construction of its IgE epitopes, are summarized in Figure 3A. Although polymorphism in the nucleotide sequence of the Asp f 3 gene was detected, no amino-acid substitutions occurred. All of the strains contained two IgE epitope sites (Figures 3A-3C).
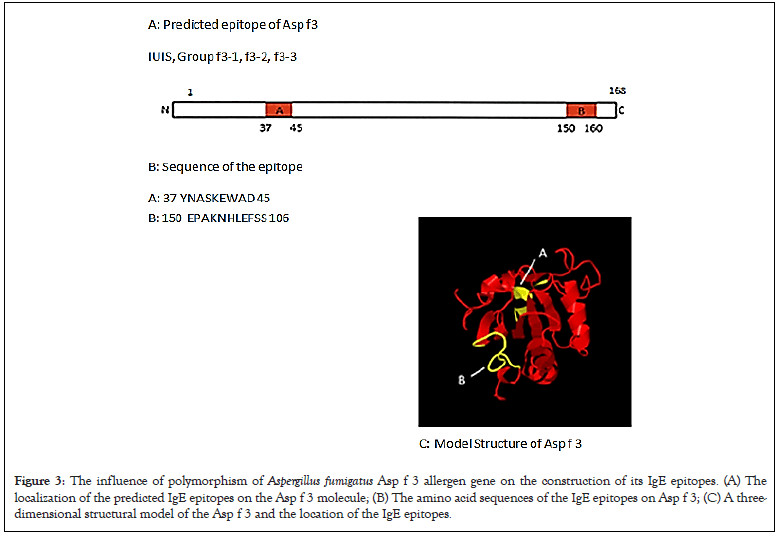
Figure 3: The influence of polymorphism of Aspergillus fumigatus Asp f 3 allergen gene on the construction of its IgE epitopes. (A) The localization of the predicted IgE epitopes on the Asp f 3 molecule; (B) The amino acid sequences of the IgE epitopes on Asp f 3; (C) A three- dimensional structural model of the Asp f 3 and the location of the IgE epitopes
Nucleotide mutations of A. fumigatus allergen genes, Asp f 1, Asp f 2, and Asp f 3, were studied. Asp f 1 gene was classified into two groups, f1-1 and f1-2 according to the mutations (Table 1). The sequence of group f1-1 was the dominant one, and was completely identical to that registered in the IUIS allergen database. A single nucleotide polymorphism of the Asp f 2 gene led to 7 different genetic groups (f2-1 to f2-7) (Table 1). Among the sequences of the Asp f 2 gene, the sequence registered in the IUIS allergen database was not found in the tested strains. The Asp f 3 gene was classified into three groups (f3-1 to f3-3) based on its nucleotide polymorphisms (Table 1). The sequence of the Asp f 3 of the f3-1 group was in complete accordance with that registered in the IUIS database. One nucleotide mutation in the f3-2 group and two mutations in the f3-3 group occurred. Categories A to G were provided, depending on the grouping of the three allergen genes of A. fumigatus (Table 2). In particular, categories A, B, and E were the major concerns. Categories A and B existed in both the environment and disease (animal/human- body) origins, suggesting no origin-specificity. However, category E was markedly specific to the environment (Table 2). Category E consisted of groups f1-1, f2-5, and f3-2. Group f1-1 was distributed into all A. fumigatus strains, and group f3-2 was recognized in strains derived from the environment and animal/human- body. However, group f2-5 was the particular one found in the environment. This fact indicates that a particular mutation of the nucleotide, depending on its origin, relates to a stable settlement to its living circumstances. It is believed that each A. fumigatus strain has adapted to its individual situation, accompanied by the mutation of some genes, such as allergen genes.
Two functional IgE epitopes were experimentally identified on the Asp f 1 molecule [12]. One of the epitopes was located from the 7th to the 22nd amino-acids from the N-terminal end. The other was located from the 140th to the 149th amino-acids [13, 14]. The Asp f 1 gene mutation described in this study resulted in an amino-acid substitution (Table 1 and Figure 1A). When an analysis was performed using computer software to predict IgE epitopes, five epitopes existed in the Asp f 1 allergen of group f1-1 (Figure 1A). The mutation in the nucleotides of group f1-2 resulted in the deletion of one epitope (Figure 1A, epitope E), but such allergological modulation is almost negligible due to the small number of isolates present in group f1-2. This implies that the nucleotide sequence of Asp f 1 registered in the IUIS allergen database is unquestionably available for use in allergen research, such as the preparation of recombinant Asp f 1 proteins.
The nucleotide sequence of Asp f 2 registered in the IUIS allergen database was not found in any of the strains tested, suggesting that the registered sequence would not be suitable for allergen research. A single nucleotide polymorphism of the Asp f 2 gene induced two allergological types. One is based on the sequence of group f2-1, but the significance of the presence of group f2-1 would be negligible because of the markedly small number of isolates present in group f2-1. The other type consists of group’s f2-2 to f2-7 (Figure 2B). The epitope site at 155 YTTRR 159 was experimentally determined [14]. This epitope might be affected by the amino-acid substitution at the 159 th position. On the other hand, the amino-acid substitution at the 297th position generated a new IgE epitope (Figure 2A, epitope E, panels B, C, E). As mentioned above, group f2-5, which possesses a newborn IgE epitope, is markedly specific to the origin of the environment. It would be possible to consider that A. fumigatus strains belonging to group f2-5 were mutated and adapted to the environment, accompanied by the acquisition of a new IgE epitope. A. fumigatus strains in group f2-5 might be more allergic than others.
The Asp f 3 genes were classified into 3 groups based on the nucleotide polymorphism. The sequence of Asp f 3 of the f3-1 group was in complete accordance with that registered in the IUIS database. One nucleotide mutation in the f3-2 group and two mutations in the f3-3 group were found. These two nucleotide substitutions did not induce any amino-acid substitutions. A conformational IgE epitope was experimentally determined in the Asp f 3 molecules [15]. The amino-acid sequence of Asp f 3 is appropriate for any kind of allergen research because there are no amino-acid substitutions.
It is known that the influenza virus mutates depending on its origin, accompanied by an increased pathogenicity [16,17]. The Asp f 2 allergen of group f2-5 possesses one more epitope, compared with the other genetic type, suggesting its facilitated pathogenicity. These findings presume that the isolates of A. fumigatus belonging to Category E would adapt to circumstances, such as the environment, via the mutations in the Asp f 2 gene. Category E was markedly specific to the environmental origin. As mentioned above, group f2-5 was strictly related to group f3- 2, which was the dominant group in Asp f 3. Although there is still no explanation for how the Asp f 2 gene of the f2-5 group relates to the Asp f 3 gene of group f3-2 described in this communication, the relationship between the adaptation of fungi to the environment and the distribution of IgE epitopes on allergic proteins is still interesting.
The polymorphism of A. fumigatus allergen genes occurred via mutations, and this polymorphism was related to the circumstances from which strains were isolated. The mutated nucleotide sequences affected their protein properties, accompanied by an IgE epitope structure. In pathogenic viruses, an increase in pathogenicity occurs due to mutations related to their origins. Our findings suggest that nucleotide mutation of fungal allergen genes, associated with the origin from which the fungus was isolated, modifies the structure of the proteins, affecting their pathogenic properties, such as their IgE epitope localization.
We thank Mrs. Yukiko Shirafuji (Iwate University) for her experimental assistance.
R.K. and Y.K. conceived and designed this study. R.K., D.I., and A.Y. performed the experiments. R.Y. and Y.K. prepared the Tables 1-3. R.Y. and Y.K. prepared Figures 1-3. All authors varied and the data. Y.K. wrote the main manuscript. All authors reviewed the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest related to the contents of this article.
Citation: Konuma R, Watanabe M, Irikura D, Sugita-Konishi Y, Yamazaki A, Yanagi U, et al. (2022) Polymorphism of Aspergillus fumigatus Major Allergen Genes Associating with Their Isolated Sites Affects Their Ige Epitope Structures. Fungal Genom Biol. 12:195.
Received: 17-Jun-2022, Manuscript No. FGB-22-17982; Editor assigned: 20-Jun-2022, Pre QC No. FGB-22-17982 (PQ); Reviewed: 07-Jul-2022, QC No. FGB-22-17982; Revised: 14-Jul-2022, Manuscript No. FGB-22-17982 (R); Published: 22-Jul-2022 , DOI: 10.35841/2165-8056.22.12.195
Copyright: © 2022 Konuma R, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.