Fungal Genomics & Biology
Open Access
ISSN: 2165-8056
ISSN: 2165-8056
Mini Review - (2021)Volume 11, Issue 5
Our previous study reported the variation of mutations in the albino strains of Aspergillus luchuensis. This short commentary builds on this previous study by further investigating the differences in mutations of the five albino strains used in that work by focusing on pigment biosynthesis. The albino strains cultured on potato dextrose agar plates showed different conidial colors, suggesting that they have different mutations in pigment biosynthesis genes. The genes targeted to investigate the mutations were obtained from public databases and previous reports on A. niger, A. fumigatus, and A. terreus, the pigment production of which has been well studied. Comparison of the amino acid sequences revealed that some mutations are shared among the albino strains, while others are unique to a specific strain. The results suggest the possibility that A. luchuensis and its albino strains can help to reveal the cryptic biosynthetic pathway behind pigment production in Aspergillus.
Albino mutant; Aspergillus luchuensis; Pigment production; Enzyme; Fermentation
Aspergillus luchuensis is an important fungus in the section Nigri for Japanese brewing culture [1]. The section Nigri has another industrially important species, A. niger, which is used globally for enzyme production and citrate fermentation. A. luchuensis has been used only in limited geographical regions, so researchers globally may have overlooked its potential as a research target and fermentation host. However, recent studies on A. luchuensis have intensified and focused on investigations of enzyme production [2-5]. Citric acid [6] and secondary metabolites [7]. Studies of A. luchuensis have often concentrated on its albino mutant, IFO4308 (=NBRC4308), because albino mutant strains are widely used in the production of “shochu,” a Japanese distilled spirit. Our previous study [8] identified the mutations of genes in albino mutants obtained from three koji starter culture manufacturers by comparing the genome of A. niger, a phylogenetically close relative of A. luchuensis, and both black and albino strains of A. luchuensis. Genome sequences of A. luchuensis are now available for strains IFO4308 [9], CBS106.47, and RIB2604 (=NBRC4314) [10]. Recently, genome sequences of strain RIB2601assembled to the chromosome level were added to a public database [11-18]. Adding the draft genomes of eight strains (TK-86–93) that we assembled ourselves, the genomes of a total of 12 strains were available for the present study. Among these genomes, seven are from black strains (CBS106.47, RIB2604, RIB2601, TK-86–89) and five are from albino strains (IFO4308, TK-90–93). This abundance of genomic data should help to describe the genomic features in each strain and reveal the physiology of A. luchuensis.
Pigment production is a good focus for describing genomic features using the accumulated genomic data of albino strains of A. luchuensis. Our previous study focused on only one gene in albino strains of this species related to pigment biosynthesis [8] but in the literature on Aspergillus research more pigment biosynthesis-related genes are proposed. This short commentary discusses the contribution of genes to conidial pigmentation by analyzing the associations of the phenotype of conidial color with mutations in genes potentially related to pigmentation and comparing them between albino and normally colored A. luchuensis strains. Comparative genomic study should help to solve the puzzle of how pigments are synthesized in Aspergillus.
Figure 1 shows that the black koji strain, TK-86, is completely black, but the “albino” strains, IFO4308 and TK-90–93, are not completely white. Furthermore, the albino strains show different conidial colors. Strain IFO4308 is relatively greenish, while TK-91 has a fawn color, but both are relatively dull and dim in color. In contrast, the other strains (TK-90, 92, 93) exhibit a bright red/orange color. This suggests a difference in pigment production among greenish IFO4308, fawn TK-91 and brightly colored TK-90, 92, and 93. A previous study showed that the pigment production mutants of A. niger also produce green or fawn conidia (Jørgensen et al.), so A. luchuensis might have a pigment production pathway similar to that of A. niger.
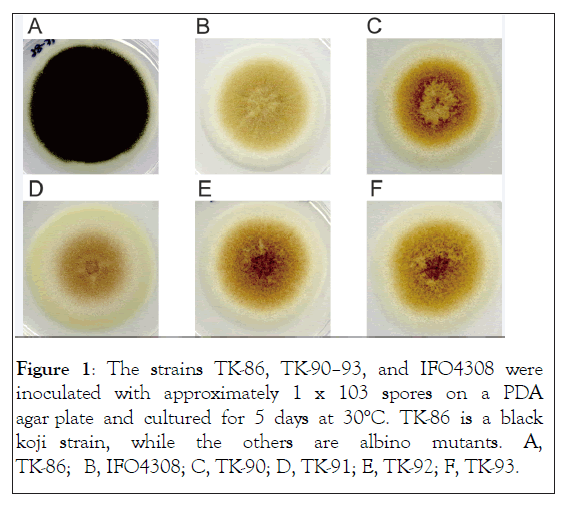
Figure 1: The strains TK-86, TK-90–93, and IFO4308 were inoculated with approximately 1 x 103 spores on a PDA agar plate and cultured for 5 days at 30°C. TK-86 is a black koji strain, while the others are albino mutants. A, TK-86; B, IFO4308; C, TK-90; D, TK-91; E, TK-92; F, TK-93.
Pigment produced by A. niger was originally identified in 1891 and named aspergillin (Ray and Eakin). Aspergillin is now recognized to consist of two different pigments: a brown one and a green one. Other pigments produced by Aspergillus species have also been studied, such as DHN-melanin (Tsai et al.) and pyomelanin in A. fumigatus (Schmaler-Ripcke et al.), and Asp-melanin in A. terreus (Geib et al.). As described in the following section, the effects of gene mutations on conidial color in the albino strains of A. luchuensis are evaluated using the results of these studies that clarified pigment biosynthetic pathways. In addition, mutations in transcription-regulating genes involved in pigment biosynthesis that were recently reported are examined. The annotations of genes were obtained from previous reports and AspGD (https://www.aspgd.org/). Table 1 shows the IDs of all of the genes evaluated in this study.
| Gene product | Gene IDs (gene name in A. niger or A. luchuensis if shown) | Reference |
|---|---|---|
| PKS | An09g05730 (fwnA) | (Jørgensen et al., 2011) |
| a/b hydrolase | An14g05350 (olvA) | (Jørgensen et al., 2011) |
| THN reductase | An16g01650, An19g00390, An07g01830, An06g01980 | (Tsai et al., 1999) |
| Scytalone dehydratase | An08g09920 | (Tsai et al., 1999) |
| Laccase | An16g02020 (mcoM), An12g05810 (mcoJ), An05g02340 (mcoF), An15g05520 (mcoK), An18g02690 (dhgo), An01g08960 (mcoH), An08g08450 (mcoG), An01g00860 (mcoN), An11g03580 (mcoD), An14g05370 (brnA) | (Tsai et al., 1999) (Jørgensen et al., 2011) |
| 4´-phosphopantetheinyl transferase | An12g03950 (ppt) | (Jørgensen et al., 2011) |
| Dioxygenase | An11g02200, An11g02180 | (Schmaler-Ripcke et al., 2009) |
| NRPS | An02g05070, An03g03520, An03g06010, An04g06260, An05g01060, An06g01300, An08g02310, An09g00520, An09g01690, An11g00050, An12g02840, An12g07230, An13g03040, An15g07530 | (Geib et al., 2016) |
| NRPS-like | An01g11770, An02g00210, An02g00840, An02g10140, An09g00450, An09g05110, An12g10860, An13g02460, An16g00600 | (Geib et al., 2016) |
| NRPS/PKS hybrid | An02g08290, An11g00250, An11g06460, An18g00520 | (Geib et al., 2016) |
| Tyrosinase | An15g07670 | (Geib et al., 2016) |
| Cytosolic protein | An11g02190 | (Keller et al., 2011) |
| Transcriptional regulator | An11g02150 | (Keller et al., 2011) |
| An11g06750 (cwcA), | (van Leeuwe et al., 2020) | |
| An01g07390 (sirD) | (Miyamoto et al., 2020) |
Table 1: Genes targeted in this study. Genes were selected from reports on A. niger or A. luchuensis or selected from AspGD (https://www.aspgd.org) using IDs of genes from A. fumigatus, or using the name of the gene product in A. terreus. PKS, polyketide synthase; THN, 1,3,6,8-tetrahydroxynaphthalene; NRPS, non-ribosomal protein synthase.
| Gene ID | TK-86 | TK-90 | TK-91 | TK-92 | TK-93 | IFO4308 |
|---|---|---|---|---|---|---|
| An09g05730 | - | E1821Cfs, V1863_P1877del, H1878P, R1879Sfs | E1821Cfs, V1863_P1877del, H1878P, R1879Sfs | E1821Cfs, V1863_P1877del, H1878P, R1879Sfs | E1821Cfs, V1863_P1877del, H1878P, R1879Sfs | V1863_P1876del, H1878P, R1879Sfs |
| An14g05350 | - | V208I | - | V208I | V208I | - |
| An16g01650 | - | - | N249_E250insGGML | - | N249_E250insGGML | N249_E250insGGML |
| An03g06010 | - | P1673H, V1899I | - | - | - | - |
| An08g02310 | - | - | K168_C169ins37 | K168_C169ins37 | - | - |
| An12g07230 | - | A1092V | A1092V | A1092V | A1092V | - |
| An15g07530 | - | A4431V | A4431V | A4431V, V4693A | A4431V, V4693A | - |
| An02g00840 | - | T111P | T111P | T111P | T111P | - |
| An04g01150 | - | - | - | - | - | M1_M6del |
| An04g04380 | - | G374D | G374D | G374D | G374D | - |
| An02g08290 | - | I2240L | I2240L | I2240L | I2240L | - |
| An11g06460 | - | I2240L | I2240L | I2240L | I2240L | - |
Table 2: Mutations in genes potentially related to pigment biosynthesis in each.
Genes involved in pigment synthesis in A. niger
When attempting to gain insights into pigment synthesis in A. luchuensis, findings obtained from A. niger should be particularly reliable because these species are phylogenetically close relatives. A previous study identified four genes related to pigment synthesis in A. niger (Jørgensen et al.): polyketide synthase (fwnA, or commonly called pskP), a/b hydrolase (olvA), laccase (brnA), and 4¢-phosphopantetheinyl transferase (ppt). Among these, fwnA, olvA, and brnA are orthologs of alb1, ayg1, and abr1 in the dihydroxynaphthalene (DHN)-melanin biosynthetic pathway in A. fumigatus, respectively. The mutants of fwnA, olvA, and brnA showed fawn, green, and brown colors, respectively (Jørgensen et al.). In addition, the ppt-deficient mutant turned white and produced no pigment. The simplest conclusion to draw from these results is that A. niger has a DHN-melanin pathway similar to that in A. fumigatus to produce green and brown pigments. However, the lack of experimental evidence means that no definitive conclusion on this issue can yet be drawn.
As we previously reported, all albino strains of A. luchuensis analyzed in this study have a mutation in the ortholog of fwnA (An09g05730) (Table 2). While the TK-90-93 strains have additional mutations in this gene compared with IFO4308. These additional mutations may influence pigment production because IFO4308 showed a different color from TK-90-93. Meanwhile, the ortholog of olvA (An14g05350) contains the same mutation in TK-90, 92, and 93 (Table 2). The conidial colors of TK-90, 92, and 93 are much brighter than those in the other A. luchuensis strains analyzed in this study. Owing to the additional mutations in fwnA in TK-90, 92, and 93, it is difficult to evaluate the effect of olvA mutation on pigment synthesis in isolation, but mutated OlvA may have broader specificity for the substrate(s) to produce several pigments. All of the albino strains showed the same amino acid sequences in the orthologs of brnA and ppt. It is reasonable that the ppt gene contains no mutations in the albino strains because their phenotypes indicate that they do actually produce some pigments.
Genes involved in pigment synthesis in A. fumigatus
Although the issue of whether A. luchuensis as well as A. niger has a DHN-melanin production pathway is still under investigation, A. luchuensis has several genes with functions known to be related to this pathway. DHN-melanin biosynthesis starts from a polyketide synthase (PKS) reaction and 1,8-DHN is the final precursor identified. The DHN-melanin biosynthetic pathway in A. fumigatus requires six genes:alb1,ayg1, abr1, arp1, arp2, and abr2. This section focuses on the last three of these, arp1, arp2, and abr2,which were not mentioned in the previous section. The ortholog of arp1 (encoding scytalone dehydratase) in A. fumigatus (An08g09920 in A. niger) has no mutation in any of the albino strains. The ortholog of arp2 (encoding 1,3,6,8-tetrahydroxynaphthalene reductase) in A. fumigatus (An16g01650 in A. niger) has the same mutation in IFO4308 and TK-91, but TK-93 also has the same mutation (Table 2). This gene may thus not have a direct effect on pigment production. No ortholog of abr2 (encoding laccase) in A. fumigatus was found in A. niger. Therefore, it is highly possible that A. luchuensis also has no ortholog of abr2. Abr2p, the product of the abr2 gene, is a potential laccase involved in the DHN-melanin production pathway (Tsai et al.). However, the lack of an ortholog of it in A. luchuensis may imply that there is no DHN-melanin production pathway in this species.
Pyomelanin and asp-melanin derived from tyrosine
The pathway of pyomelanin, a derivative of tyrosine, has been well studied using A. fumigatus. The genes related to pyomelanin production are shown in Table 1. There are no mutations in these genes in the entire albino strains used in this study, so the pyomelanin pathway does not contribute to the difference in pigment synthesis among these albino strains.
The other pigment derived from tyrosine is Asp-melanin, which is assumed to be specific to A. terreus. It is proposed that tyrosinase, non-ribosomal protein synthase (NRPS), and NRPS-like enzyme are related to the production of Asp-melanin. The ortholog annotated as tyrosinase in A. niger (An15g07670) has no mutation in any of the albino strains. There are 37 genes annotated as encoding NRPS and NRPS-like enzymes, including NRPS/PKS hybrid enzyme. Several mutations were found in the genes related to Asp-melanin production Table 2, but most of them were shared among TK-90–93 strains, so they do not correspond to the phenotype of pigment production. Some genes have strain-specific mutations, such as NRPS-like enzyme (An04g01150) in IFO4308, NRPS (An03g06011) in TK-90, and another NRPS (An08g02310) in TK-91 and TK-92. IFO4308-specific mutation may thus be related to pigment biosynthesis.
Genes regulating pigment synthesis
A recent study reported that sirD in a sirtuin biosynthetic gene cluster may be related to pigment biosynthesis in A. luchuensis (Miyamoto et al.). Specifically, a mutant with sirD deletion showed paler mycelial color than the wild type. Another study reported that cwcA may act as a negative regulator of cw1 in A. niger (Leeuwe et al.). Specifically, a mutant with cwcA deletion showed the production of an unknown yellow pigment. However, all albino strains in this study have wild-type sirD (An01g07390) and cwcA (An11g06750).
This paper provides detailed descriptions of the mutations in pigment-related genes in albino strains of A. luchuensis in order to evaluate their effects on pigment biosynthesis in this species. In several genes, mutations were shared among the albino mutant strains. Intriguingly, the olvA (An14g05350) gene has a mutation in the albino strains exhibiting a relatively strong conidial color, although the reason for this remains unclear. This mutation may change the specificity of substrate to establish a novel pigment biosynthetic pathway. It should also be noted that several NRPS genes have mutations in a specific albino mutant. These NRPS genes may be related to a pigment biosynthetic pathway. These results can help to identify the unknown pathway(s) for pigment biosynthesis in A. luchuensis and further reveal similar pathways in A. niger. Comparative study of various strains including black and albino strains of A. luchuensis using genomic data is still at an early stage. However, A. luchuensis could be a new frontier in the study of fungal genomics and physiology.
We thank Edanz (https://jp.edanz.com/ac) for editing the English text of a draft of this manuscript.
Citation: Sawada K, Yamada T (2021) Mutations in Genes Involved in Pigment Synthesis Pathways in Albino Mutants of Aspergillus luchuensis. Fungal Genom Biol.11:174.
Received: 19-Nov-2021 Accepted: 03-Dec-2021 Published: 10-Dec-2021
Copyright: © 2021 Sawada K, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.