Poultry, Fisheries & Wildlife Sciences
Open Access
ISSN: 2375-446X
ISSN: 2375-446X
Research Article - (2023)Volume 11, Issue 3
Adults of many bird species exhibit morphological distinctions between sexes. However, sex determination of monomorphic bird species on the basis of physical appearance is difficult. Molecular sexing with an appropriate primer pair is widely adopted to determine the sex of monomorphic birds. Sarus crane is one such endangered monomorphic bird whose sex can be identified using P2/ZW common primer pair or P2/W specific primer pair. We identified the sex of a Sarus crane rescued at CNP through molecular sexing, based on size differences between introns of the CHD gene on the Z and W sex chromosomes. The rescued bird was identified as male and sent to central zoo for pairing with an already existing female there. This was a very crucial step in avian wildlife conservation and for captive breeding management in the zoo. PCR based sex identification is an easy, accurate, sensitive, feasible, and highly approved method to perform molecular sexing of the rescued monomorphic birds.
Sarus crane; Molecular sexing; CHD gene; Polymerase chain reaction; Captive breeding
In the case of monomorphic birds, it is not possible to identify the sex on the basis of visual observation because males and females have similar phenotypic traits regardless of their differences in reproductive systems. For reliability and rapidity of sex identification in birds exhibiting sexual monomorphism, the molecular sexing technique was developed and the technique is continuously improving. Molecular sexing is critical for the management and conservation of avian wildlife. Accurate sex identification in birds is essential in the management of population, behavioral and ecological studies, as well as in the improvement of ex situ captive breeding programs. The molecular technique of sex identification based on PCR is a standard method for sex identification. Sarus crane is one of the monomorphic birds with a dwindling population due to the rapid deterioration of their habitats, especially wetlands, leading the population to the verge of extinction [1].
It is important to identify sex reliably to achieve a balanced sex ratio for avian wildlife management and captive breeding program. In birds, contrary in mammals, the females are heterogametic sex (ZW) while males are homogametic sex (ZZ) and the bird sex chromosomes have evolved from a pair of autosomes. During evolution, the W chromosome lost many of its genes, so its size was reduced, while the Z chromosome has not changed its gene content. The sex chromosomes are distinguished from one another on the basis of their size difference; the Z chromosome falls into the group of large chromosomes, called macro chromosomes while the W chromosome represents smaller chromosomes, called micro chromosomes (Figure 1) [2].
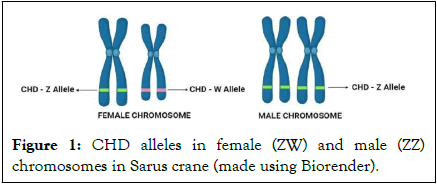
Figure 1: CHD alleles in female (ZW) and male (ZZ) chromosomes in Sarus crane (made using Biorender).
A highly conserved CHD gene that encodes chromodomain helicase DNA binding protein has distinct gametologues in the Z (CHD-Z) and W (CHD-W) chromosomes which differs in the fragment size of the intronic regions and lacks autosomal copies or pseudogenes. The most widely used molecular methods for sexing birds use the size differences between introns of the CHD gene on the Z and W sex chromosomes. Heterogametic (ZW) females are expected to have two different sized introns with 153 bp CHD-Z and 263 bp CHD-W while homogametic (ZZ) males have one specific sized intron with 153 bp CHD-Z. Consequently, ZZ is identified by 1 band and ZW by 2 bands on a gel with P2/ZW-common primer pair and ZZ is identified by no band and ZW by 1 band on a gel with P2/W-specific primer pair [3].
In our study, molecular sexing of an injured Sarus crane rescued from the CNP was conducted before it was taken to the Jawalakhel zoo at Lalitpur, Nepal for the purpose of sex identification required for captive breeding. The molecular sexing was carried out with the aid of qualitative PCR [4]. Once the positive male or female band is identified, all the other rescued birds can be compared with that positive sample if needed in the future.
Sample collection
The blood sample was collected from the jugular vein of the Sarus crane by the veterinary team from NTNC Biodiversity Conservation Center (NTNC-BCC) who were involved in the rescue mission led by the government authorities from CNP. The blood samples were collected in an Ethylene Diaminetetraacetic Acid (EDTA) buffer containing tube. The DNA was extracted immediately after it was collected in the NTNC-BCC molecular lab [5].
DNA extraction
Small part of the fresh blood sample was used for DNA extraction and the remaining sample was stored at -4°C for future use. DNA was extracted by using the DNeasy Blood and Tissue Kit (QIAGEN) following the manufacturer's protocol. The cell was lysed using proteinase K and lysis buffer. The DNA was collected in a 200 μl elution buffer. The DNA sample was stored at -20°C for future use. The extraction was carried out in a sterile and contamination free environment. All the reagents and equipment used were sterilized and the work surfaces were kept clean using 70% ethanol [6].
Polymerase Chain Reaction (PCR)
Qualitative PCR was carried out using a Bio-Rad T100 TM thermal cycler. PCR reaction was carried out in 20 μl reaction volume containing 10 μl My Taq Red Mix (2X) from Bioline, 1 μl forward primer, 1 μl reverse primer, 3 μl template DNA, and 4 μl nuclease free water. In the PCR negative control, no DNA template was added; instead, 8 μl nuclease free water was added. One negative PCR control for each primer pair was used. The amplification reaction was carried out as follows: Initial denaturation at 94°C for 10 min, followed by 45 cycles of denaturation at 94°C for 30 sec, annealing at 56°C for 30 sec., extension at 72°C for 20 sec, and final extension at 72°C for 5 min [7].
| Primers | Sequence | Melting temperature, Tm |
|---|---|---|
| P2 | 5'-TCTGCATCGCTAAATCCTTT | 61.3°C |
| W-specific | 5'-GTTTTCACACATGGCACA | 59.0°C |
| ZW-common | 5'-GATCAGCTTTAATGGACTGAAG | 59.4°C |
Table 1: Primer pairs used: P2/W-specific and P2/ZW-common.
Once the positive male sample was confirmed from this experiment, it was further used as a positive control to perform molecular sexing of other unknown samples which are suspected as male [8].
The PCR products were run in 2% agarose gel stained with EtBr. A 100 bp to 2000 bp Easyladder I from Bioline was used to compare the band of the Z or W chromosome. The bands were visualized under UV light in the GeNeiTM gel documentation system [9].
First, the Sarus crane was rescued and safely treated by a team of veterinarians as it was naturally injured. Second, the sex was identified and the bird was transported to the central zoo to restore its health and for captive breeding. Then, it was paired up with another female Sarus crane in the zoo. It is not safe for the Sarus to be sent back into nature on its own before it is healthy due to the risk of predators [10].
In this experiment, we did not observe any visible bands in both the PCR controls (Wc and ZWc) which suggests that the experiment was carried out without any cross contamination. No band was observed with the P2/W specific primer pair because W specific primer does not have a binding site on the CHD-Z allele in the case of a male. However, one band was observed with P2/ZW-common primer pair as ZW common primer has a binding site on the CHD-Z allele for males (Figure 2) [11].
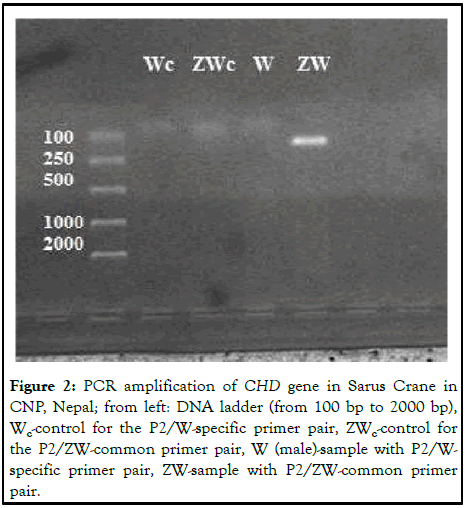
Figure 2: PCR amplification of CHD gene in Sarus Crane in CNP, Nepal; from left: DNA ladder (from 100 bp to 2000 bp), Wc-control for the P2/W-specific primer pair, ZWc-control for the P2/ZW-common primer pair, W (male)-sample with P2/W-specific primer pair, ZW-sample with P2/ZW-common primer pair.
Hence, after the PCR and gel electrophoresis of the DNA extracted from the blood sample, the Sarus was identified as male. The PCR technique can be used to identify the sex of monomorphic birds with the right combination of primer pairs. This positive result for males was used as a positive control to sexually identify another rescued male Sarus crane (Figure 3) [12].
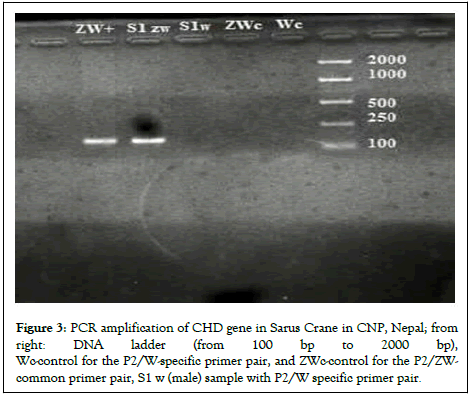
Figure 3: PCR amplification of CHD gene in Sarus Crane in CNP, Nepal; from right: DNA ladder (from 100 bp to 2000 bp), Wc-control for the P2/W-specific primer pair, and ZWc-control for the P2/ZW-common primer pair, S1 w (male) sample with P2/W specific primer pair.
We successfully applied the molecular sexing technique to identify rescued Sarus crane. Whether the birds are being managed in situ or ex-situ, it is essential to accurately identify their sex so as to maintain a proper sex ratio balance [13]. In our experiment, we used P2/ZW common primer pair and the P2/W specific primer pair to identify the sex of a rescued Sarus crane. Through the molecular sexing process, the Sarus crane was identified as a male since only one band was observed at ~153 bp.
Population sex ratio is often male biased in monogamous birds; one of the suspected reasons is sex biased predation. Sex identification in avian species is a cardinal step in avian breeding and evolutionary studies. Gender assay in a wide range of bird species can be done using the primer sets related to CHD genes. The CHD-W gene is located on the W chromosome; therefore it is unique to females whereas the other gene, CHD-Z, is found on the Z chromosome and therefore occurs in both sexes (female, ZW; male, ZZ). According to Insee, et al., P2/P8 primer pair, even though extensively used for diverse bird species for a very long time period, has certain challenges in sex identification due to a very small size difference between CHD-Z and CHD-W fragments. For the intronic length based method, Insee, et al., found that the difference in size between CHD-Z and CHD-W fragments is 170 bp, and hence could be identified using an agarose gel. The difference in size between CHD-Z and CHD-W fragments amplified with the same primer set for some other bird’s ranges from 150 bp to 250 bp. However, P2/P8 is not preferred for a few bird species. P2/ZW common and P2/W specific primer pairs work really well for Sarus crane molecular sexing. The increased amplicon size difference makes it easy to identify the different sex using PCR based method followed by simple gel electrophoresis [14-18].
To sum up, the molecular sexing technique comes in handy and can be applied in the case of monomorphic birds during the rescue process. It does not even require any restriction enzymes or cloning vectors. Knowing the right combination of primers on the basis of the gene size difference is crucial in molecular sex identification techniques [19-21].
In our study, we applied molecular sexing in birds during the rescue mission and highlighted the conservation implications. The monomorphic birds extricated from serious wild circumstances can be sexually identified through molecular sexing methods using the right combination of forward and reverse primers. After identifying its sex as male, the Sarus was sent to the zoo and paired up with the female. This is an important step in captive breeding management. This method can also be used to identify the sex of the dead birds in the wild. PCR based sex identification is crucial in both in situ and ex situ maintenance of monomorphic birds.
We would like to acknowledge the DNPWC for permitting this research. The experiments comply with the current laws of the Nepal government. The editors reserve the right to reject manuscripts that do not comply with this requirement. The research was funded and carried out in the NTNC-BCC molecular biology lab during the rescue of Sarus crane. Hence, we would like to extend our hearty gratitude to NTNC-BCC for facilitating this research and most importantly Mr. Ram Kumar Aryal, the then NTNC-BCC officer in-charge. We would also like to extend our heartfelt thanks to the technicians and caretakers of Sarus crane.
All authors contributed to the study, conception, and design. Sample collection, molecular sexing, and result interpretation were performed by Prakriti Kandel, Janardan Dev Joshi, Lasta Shrestha, Amir Sadaula and Babu Ram Lamichhane. The first draft of the manuscript was written by Prakriti Kandel and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Not required because the experiment was under the policy and guidance of Nepal government.
The study was permitted by DNPWC. This study was financially supported by NTNC-BCC and no other grants were received. There are no conflicts of interest for this work.
The authors have no competing interests to declare that are relevant to the content of this article.
All data generated or analyzed during this study are included in this article.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Kandel P, Joshi JD, Shrestha L, Sadaula A, Lamichhane BR, Upadhyaya SK, et al. (2023) Molecular Sexing of Sarus Crane (Grus antigone) Using Polymerase Chain Reaction (PCR) Technique in Chitwan National Park (CNP), Nepal and Its Conservation Implications. Poult Fish Wild Sci. 11:244.
Received: 25-Apr-2023, Manuscript No. PFW-23-23744; Editor assigned: 27-Apr-2023, Pre QC No. PFW-23-23744 (PQ); Reviewed: 11-May-2023, QC No. PFW-23-23744; Revised: 30-Jun-2023, Manuscript No. PFW-23-23744 (R); Published: 07-Jul-2023 , DOI: 10.35248/2375-446X.23.11.244
Copyright: © 2023 Kandel P, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.