Mycobacterial Diseases
Open Access
ISSN: 2161-1068
ISSN: 2161-1068
Review Article - (2018) Volume 8, Issue 3
Keywords: Molecular epidemiology; Strain typing; Morocco; IS6110-RFLP; Spoligotyping; MIRUs-VNTR
TB: Tuberculosis; MTB: Mycobacterium tuberculosis; MTBC: Mycobacterium tuberculosisComplex; MIRUs- VNTR: Mycobacterial Interspersed Repetitive Units-Variable Number of Tandem Repeats; RFLP: Restriction Fragment Length Polymorphism; SITs: Shared International Types
Tuberculosis (TB), caused by M. tuberculosis (MTB), remains a major global health concern and ranks as the second leading cause of death from infectious diseases worldwide [1]. The TB surveillance is generally performed by conventional methods based on demographic and clinical information, microbiological confirmation tests and recently molecular commercial assays. The development of molecular epidemiology in recent years has revolutionized our understanding of TB transmission since it has complemented conventional epidemiologic methods in the investigation of outbreaks, nosocomial transmission, laboratory cross-contamination, determination of the proportions of TB disease attributable to recent transmission versus reactivation and to outline risk factors for recent acquisition of MTB infection and TB disease [2].
These approaches have been proven powerful tools to assess and explain the extent of genetic diversity of MTB strains throughout the world, and to shed light on the evolutionary forces that shape this diversity. There is no doubt that the immense diversity of MTB strains is linked to ancient human migrations out of Africa, as well as to more recent movements mainly to Europe during the past few hundred years. Indeed, many strains/lineages were initially endemic within specific geographical areas, and have become epidemic, sporadic or ubiquitous [3]. Moreover, the evolutionary characteristics of MTB strains are synergizing with the effects of increasing globalization and intensifying human travel and migration [4].
Despite the efforts made by the National program to fight against TB to decrease the incidence and mortality rate in the last decades as well as to limit spread of MTB strains, TB is still one major public health concern in Morocco. Epidemiological investigations have always been limited to classical epidemiology which deals with national and regional incidence/prevalence and demographic characteristics. There were a few sporadic studies that have characterize MTB strains circulating in Morocco based on one or a combination of molecular markers/methods such as RFLP (Restriction Fragment Length Polymorphism) which bases on the monitoring the number of insertion sequence IS6110 in the chromosome which varies among different strains [5], spoligotyping which detects polymorphisms present in a direct repeat (DR) locus [6] and MIRUs-VNTR (Mycobacterial Interspersed Repetitive Units- Variable Number of Tandem Repeats) (molecular epidemiology) that relies on measuring repetitive DNA elements [7]. The present paper aims to provide an overview and a longitudinal analysis of circulating M. tuberculosis Complex (MTBC) genotypes in Morocco and their transmission dynamics within the population during last decades.
A literature review was conducted by the authors using electronic databases, including Pubmed/Medline, Science Direct, Google, and Google Scholar to search for articles already published with no limitation to the language and year of publication. The terms used for the information retrieval were M. tuberculosis, Morocco, Strain typing, Molecular epidemiology, IS6110-RFLP and MIRUs-VNTR and Spoligotyping. The relevant epidemiological data were identified starting with the year 1997 to December 2017.
Three important parameters/rates namely clustering rate, recent transmission rate and the Hunter and Gaston Discriminatory Index were retrieved whenever it is possible, they are calculated by the following formula [8,9]:
Clustering rate=(nc-c)/n
Where nc is the total number of clustered isolates, c is the number of isolate clusters, and n is the total number of isolates in the sample.
Transmission rate=[Tc-Nc]/Ta*100%
Where Tc was the total number of clustered isolates, Nc was the number of clusters, and Ta was the total number of isolates
The discrimination of the locus combination is calculated using the Hunter-Gaston discriminatory index (HDGI):
HGDI=1-1N (N-1)Σj=1snj (nj-1)
Where N is the total number of isolates in the typing method, s is the number of distinct patterns discriminated by MIRU-VNTR, and nj is the number of isolates belonging to the jth pattern.
The screening of articles was performed based on the relevance of the title, abstract and manuscript review. In order to minimize the risk of bias, the data was extracted from the selected studies and inserted into a data sheet by one reviewer and across studies.
This review was conducted on studies published on genetic diversity of M. tuberculosis isolates from different regions of the Moroccan population. After the evaluations of all articles, all of them were relevant (those published from 1997 to December 2017) and thus were selected for analysis.
MTB population structure based on IS6110 RFLP
Earlier TB molecular epidemiological investigations in Morocco were based on the IS6110 RFLP analysis (Two investigations). This technique is based on the variability of the number of copies of IS6110 within MTBC genome and was long recognized as the gold standard for MTBC strain differentiation as it provides the highest discriminatory power of all available MTBC genotyping methods [5].
El Baghdadi et al. [10] used the IS6110 to differentiate 22 clinical MTB isolates from Casablanca, Morocco. The results demonstrated that IS6110 marker is a useful tool for the epidemiological survey of TB in Morocco since it has permitted to detect (i) strains with similar IS6110 RFLP patterns in 62.5% of patients confirming that they derived from same sources, (ii) identical patterns in one patient despite MTB compartmentalization (sputum and lymph node). Four sequential strains isolated from the same patients before and after one year treatment had identical IS6110 RFLP patterns suggesting relapse rather than reinfection.
A second study was conducted by Diraa et al. [11] on 61 isolates recovered from new cases of pulmonary TB in Casablanca by applying IS6110 RFLP. The results showed that most of MTB isolates (92%) harbored 6-14 copies of IS6110 meaning that IS6110 RFLP is a suitable method to type MTBC strains. However, the recent transmission rate was lower than expected in such high incidence TB community.
Overall, the information on MTB genotype diversity in Morocco, based on IS6110, is scarce but mirrors an unexpected low level of recent transmission, and a no correlation between drug resistance profiles and IS6110 RFLP patterns. These studies were very informative but would be more relevant if conducted on a higher numbers of isolates from many cities in Morocco and recovered during a longer recruitment period.
MTB population structure based on Spoligotyping data
Overall, four studies conducted using spoligotyping were included in this review. First, Brudey et al. [12] published the fourth international spoligotyping database, SpolDB4, which describes 1939 Shared International Types (SITs) representative of a total of 39,295 strains from 122 countries including 127 strains from Casablanca, Morocco. The molecular analysis namely spoligotyping has permitted to draw the first picture of the current MTBC genomes diversity from Morocco.
Similarly, Chaoui et al. [13] addressed the question of MTBC genetic diversity by typing a larger panel of M. tuberculosis isolates (n=219 corresponding to 208 patients) collected over a period of three years; between January 2002 and December 2004; the patients came essentially from cities located in west and north Morocco (Rabat, Kenitra and Tanger regrouped 87.5% of isolates), with following drug resistance/susceptible status: 49% strains were pansusceptible, 26.9% of the strains being MDR; and 24.1% with other resistance(s) profile. Of note, this sample collection was marked by a high relapse rate since 57.94% of patients with known clinical status relapsed.
Nonetheless, knowledge on MTBC population structure remained fairly understood at the national level, thus Lahlou et al. [14] focused on MTBC genotypic population structure by applying spoligotyping on 592 MTBC isolates; originated from 23 cities in Morocco.
Much later, the study conducted by Bouklata et al. [15] has permitted to track and monitor the extent of genetic diversity of MTB strains. Within this context, a total of 168 MTBC isolates were collected during 2010-2012 from 10 different Moroccan cities, and subjected to spoligotyping.
All the findings were consistent; the genetic family that dominated was the Latin American Mediterranean (LAM) (50.6%) followed by Haarlem (22.3%) and T (20.7%). Of note, each of the three families was characterized by one predominant SIT corresponding to the « prototype » namely SIT42 for LAM, SIT50 for Haarlem and SIT53 for T.
The predominance of some SITs, in particular those of LAM family, is likely to be attributed to the introduction of the corresponding MTBC clonal branches in the region (founder effect). The SIT42/LAM9 was prevalent in all the regions of Morocco, and its predominance is most likely attributed to relatively high transmission rate observed. Strikingly, its increased transmissibility within our population along with its stable association with respective human population might be the driven force of the evolution and omnipresence of such SIT.
The LAM family has been described prevalent in countries in the Americas and Europe [16]. The MTB strains currently circulating in Morocco are dominated by LAM lineage with its 6 out of 12 reported sub-lineages throughout the world [17]. Hence, more investigations based on Whole Genome Sequencing are of great value to decipher the biogeographical structure and phylogenetic history of this efficient and successful genogroup/family.
The S family is likely to be quite old (4500 years); it was first described in Sicily and Sardinia, with specific and rare shared types that suggested local microevolution and adaptation namely ST295 (Haiti), ST1334 (South Africa), ST1063 (Algeria) and ST1068 (Morocco). The latter has recently been found associated with MDR status (unpublished data), hence its presence in Morocco deserves more investigations.
Beijing is the best and the most studied MTBC lineage. First, it was introduced in African continent through the sea route from the southern sea-ports of Hong Kong, Singapore and Vietnam to South Africa [18]. Later on, this lineage was increasingly been described in sub-Saharan Africa albeit at small rates as well as in Morocco (1.1%) which reflects the low level of human immigration from East Asia, where this strain lineage largely prevails and probably originally emerged [19]. MTBC isolates with a Beijing genotype are often, albeit not always associated with MDR-TB, especially in Eurasia and East-Asian countries [20,21]. There was very little association of Beijing genotypes with MDR-TB in Morocco. However, the possible origins of the Beijing isolates identified deserve more vigorous investigations to address this issue.
From a phylogenetical window, evolutionary-modern MTBC lineages belonging to the “evolutionary recent” TbD1-/Principal Genetic Groups 2/3 TB are almost responsible of TB in Morocco.
Four studies have used spoligotyping for genetic diversity analysis (n=1055); a description of the SITs reported in Morocco so far are listed in Table 1.
| SIT | Clade | Spoligotype Description | Octal Number | Total Strains |
|---|---|---|---|---|
| 1 | Beijing | 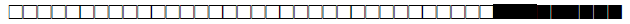 |
000000000003771 | 11 |
| 2 | Haarlem2 | 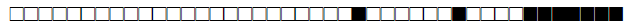 |
000000004020771 | 6 |
| 3 | H3 | 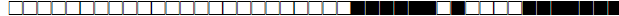 |
000000007720771 | 1 |
| 4 | LAM3 | 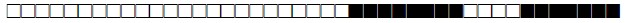 |
000000007760771 | 1 |
| 7 | T1 | 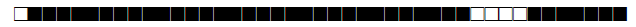 |
377777777760771 | 1 |
| 26 | CAS1-Delhi | 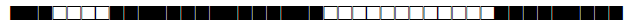 |
703777740033771 | 1 |
| 31 | T1 | 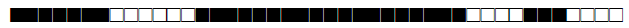 |
774037777760700 | 3 |
| 32 | U (likely S) | 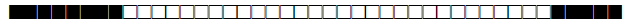 |
776000000000171 | 1 |
| 33 | LAM3 | 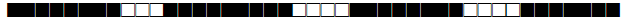 |
776177607760771 | 27 |
| 34 | S |  |
776377777760771 | 18 |
| 36 | Haarlem3 |  |
777737777760771 | 6 |
| 37 | T3 |  |
777737777760771 | 3 |
| 40 | T4 |  |
777777377760771 | 2 |
| 42 | LAM9 | 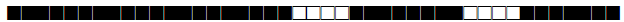 |
777777607760771 | 285 |
| 44 | T5 | 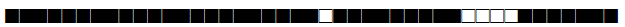 |
777777757760771 | 4 |
| 46 | U (likely H) |  |
777777770000000 | 1 |
| 47 | Haarlem1 | 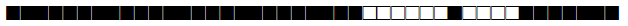 |
777777774020771 | 51 |
| 49 | Haarlem3 | 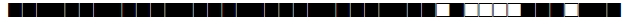 |
777777777720731 | 1 |
| 50 | Haarlem3 | 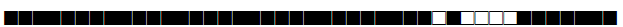 |
777777777720771 | 80 |
| 52 | T1 |  |
777777777760731 | 1 |
| 53 | T1 | 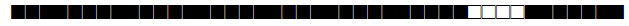 |
777777777760771 | 115 |
| 58 | T5-Madrid2 | 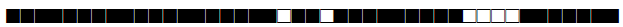 |
777777557760771 | 6 |
| 60 | LAM4 | 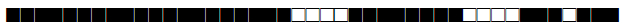 |
777777607760731 | 19 |
| 61 | LAM10-CAM | 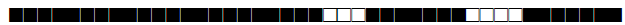 |
777777743760771 | 11 |
| 62 | Haarlem 1 | 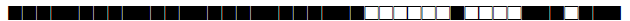 |
777777774020731 | 9 |
| 64 | LAM6 | 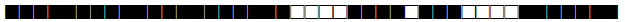 |
777777607560771 | 4 |
| 65 | T1 | 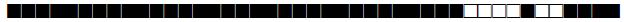 |
777777777760471 | 1 |
| 71 | S | 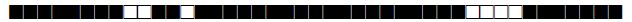 |
776337777760771 | 1 |
| 73 | T2-T3 | 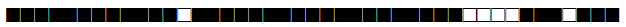 |
777737777760731 | 1 |
| 77 | T1 | 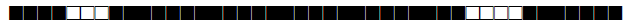 |
743777777760771 | 2 |
| 78 | T1-T2 | 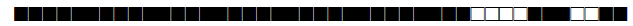 |
777777777760711 | 2 |
| 93 | LAM5 | 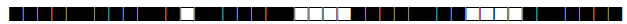 |
777737607760771 | 8 |
| 102 | T1 | 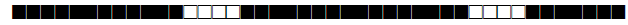 |
777703777760771 | 2 |
| 105 | LAM3 | 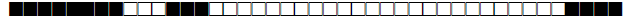 |
776160000000071 | 2 |
| 106 | LAM3 | 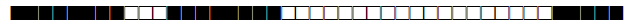 |
776177400000171 | 4 |
| 120 | T1 | 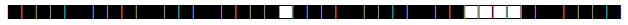 |
777777577760771 | 1 |
| 143 | Haarlem | 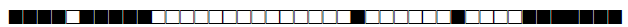 |
757400004020771 | 5 |
| 150 | LAM9 | 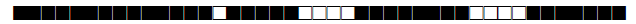 |
777767607760771 | 52 |
| 151 | Haarlem | 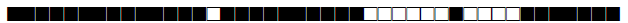 |
777767774020771 | 1 |
| 161 | LAM9 | 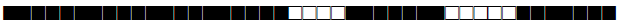 |
777777607740771 | 1 |
| 162 | LAM9 | 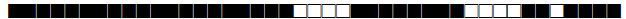 |
777777607760671 | 1 |
| 163 | LAM4 | 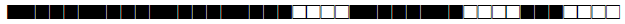 |
777777607760700 | 2 |
| 164 | T |  |
777777760060771 | 3 |
| 177 | LAM | 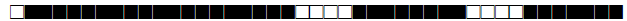 |
377777607760771 | 3 |
| 180 | Haarlem | 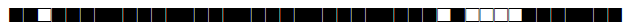 |
677777777720771 | 1 |
| 183 | Haarlem | 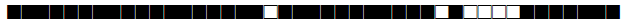 |
777777377720771 | 2 |
| 205 | T1 |  |
737777777760771 | 1 |
| 209 | LAM12- Madrid1 | 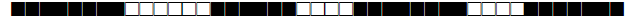 |
776017607760771 | 1 |
| 218 | H1 | 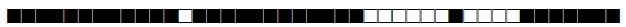 |
777737774020771 | 2 |
| 244 | T1 | 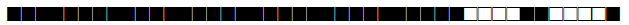 |
777777777760601 | 1 |
| 251 | T1 | 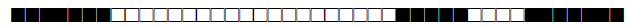 |
077400000076771 | 3 |
| 252 | LAM9 | 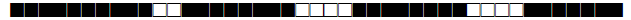 |
777477607760771 | 1 |
| 268 | H3 | 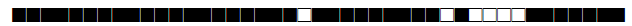 |
777777677720771 | 1 |
| 273 | LAM9 | 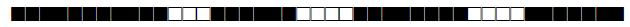 |
777617607760771 | 1 |
| 291 | T1 | 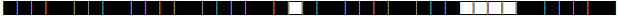 |
777777677760771 | 3 |
| 334 | T1 |  |
577777777760771 | 1 |
| 373 | T1 | 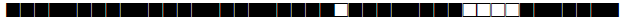 |
777777767760771 | 1 |
| 388 | LAM9 | 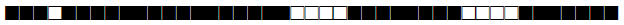 |
737777607760771 | 1 |
| 390 | H3 | 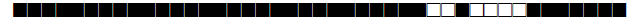 |
777777777620771 | 1 |
| 433 | H3 | 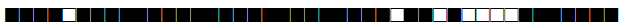 |
000000757777777 | 12 |
| 443 | U | 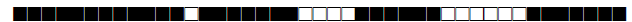 |
777737607700771 | 1 |
| 451 | H37Rv | 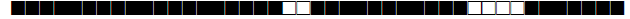 |
777777477760771 | 1 |
| 462 | T1 | 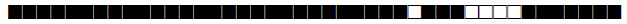 |
777777777560771 | 1 |
| 466 | S |  |
776377377760771 | 4 |
| 481 | BOV1 | 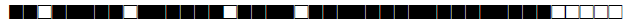 |
676773677777600 | 1 |
| 492 | LAM9 | 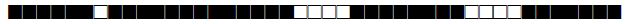 |
773777607760771 | 2 |
| 535 | T1 | 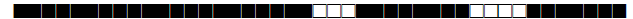 |
777777707760771 | 1 |
| 589 | U | 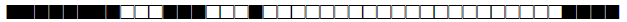 |
776161000000071 | 2 |
| 602 | U | 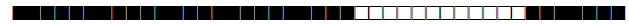 |
777777770000771 | 8 |
| 609 | H1 | 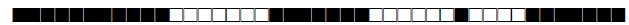 |
777600774020771 | 2 |
| 615 | H3 | 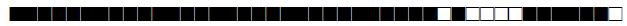 |
777777777720770 | 1 |
| 644 | BOV4-CAPRAE | 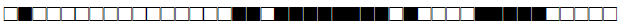 |
200003377207600 | 1 |
| 731 | LAM9 | 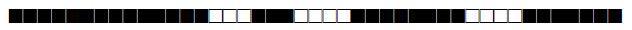 |
777761607760771 | 8 |
| 737 | LAM9 | 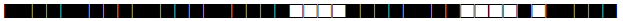 |
777777607760571 | 2 |
| 741 | H3 | 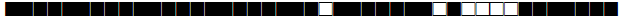 |
777777757720771 | 8 |
| 746 | H3 |  |
777777777520771 | 3 |
| 751 | T1 |  |
077777777760771 | 2 |
| 760 | H3 | 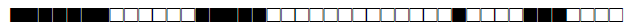 |
774037000020700 | 2 |
| 770 | LAM9 | 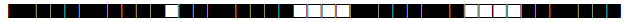 |
777677607760771 | 1 |
| 784 | T2-S |  |
776377777760731 | 5 |
| 822 | LAM9 | 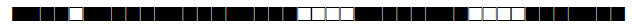 |
757777607760771 | 3 |
| 866 | LAM9 | 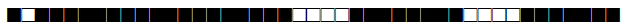 |
577777607760771 | 6 |
| 926 | T1 | 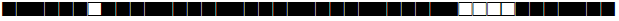 |
773777777760771 | 1 |
| 964 | LAM9 | 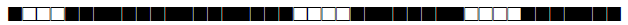 |
437777607760771 | 9 |
| 1026 | BOV1 | 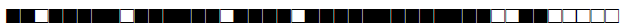 |
676773677774600 | 1 |
| 1058 | T2 |  |
037777777760731 | 1 |
| 1064 | LAM9 | 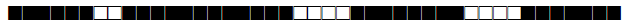 |
771777607760771 | 11 |
| 1067 | T1 | 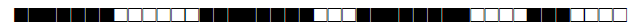 |
774037707760700 | 1 |
| 1068 | S | 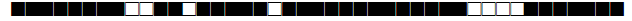 |
776337377760771 | 4 |
| 1069 | T1 |  |
771777777760771 | 8 |
| 1070 | U | 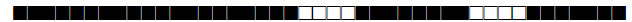 |
777777607760371 | 10 |
| 1071 | LAM9 | 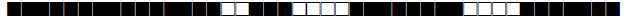 |
777771607760771 | 7 |
| 1072 | LAM5 | 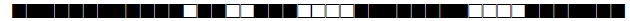 |
777731607760771 | 6 |
| 1073 | T1 |  |
777637777760771 | 1 |
| 1074 | LAM9 | 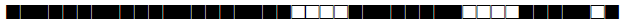 |
777777607760761 | 16 |
| 1075 | LAM9 | 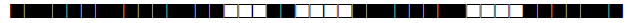 |
777770607760771 | 2 |
| 1105 | T1 | 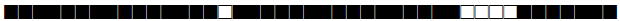 |
777773777760771 | 2 |
| 1111 | T1 | 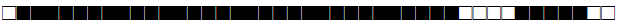 |
377777777760760 | 1 |
| 1129 | T1 |  |
776777777760771 | 1 |
| 1135 | H3 | 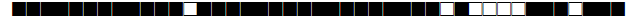 |
777737777720731 | 3 |
| 1139 | H1 | 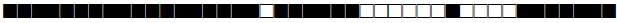 |
777777374020771 | 1 |
| 1154 | LAM9 | 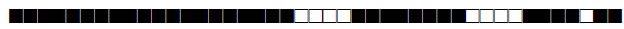 |
777777607760751 | 1 |
| 1155 | H1 |  |
577777774020771 | 1 |
| 1227 | T5-Madrid2 | 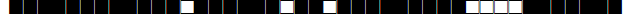 |
777737557760771 | 1 |
| 1277 | LAM9 | 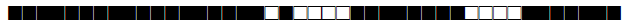 |
777777207760771 | 1 |
| 1528 | LAM9 | 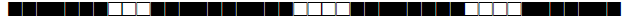 |
774377607760771 | 1 |
| 1536 | S | 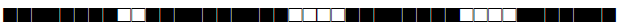 |
776377607760771 | 7 |
| 1537 | LAM3 | 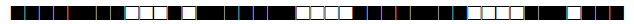 |
776137607760731 | 8 |
| 1538 | H3 | 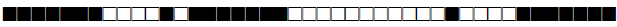 |
774137600020771 | 6 |
| 1539 | H3 |  |
773777777720771 | 1 |
| 1541 | LAM9-S | 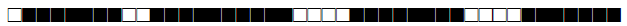 |
376377607760771 | 1 |
| 1557 | H1 |  |
777677774020771 | 1 |
| 1580 | T1 | 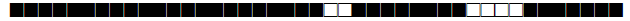 |
777777747760771 | 2 |
| 1626 | T1 | 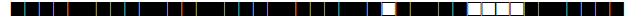 |
777777776760771 | 1 |
| 1655 | T3 | 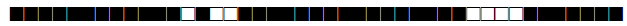 |
777723777760771 | 4 |
| 1708 | LAM9 | 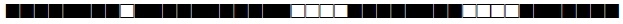 |
776777607760771 | 1 |
| 1735 | H3 |  |
777777774320771 | 1 |
| 1743 | H3 |  |
767777777720631 | 1 |
| 1778 | U | 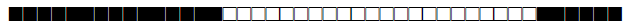 |
777770000000371 | 1 |
| 1803 | LAM9 | 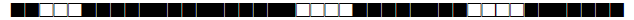 |
617777607760771 | 1 |
| 1832 | LAM9 | 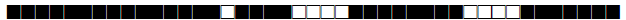 |
777773607760771 | 2 |
| 1844 | LAM9 | 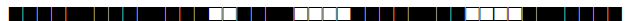 |
777763607760771 | 2 |
| 1891 | H3 | 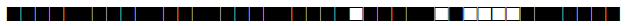 |
777777773720771 | 1 |
| 1952 | Harleem | 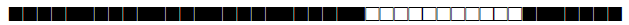 |
777777774000771 | 1 |
| 1999 | LAM5 | 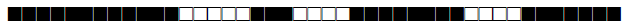 |
777701607760771 | 1 |
| 2025 | H3-T1 | 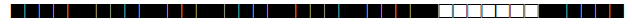 |
777777777700371 | 1 |
| 2041 | UNK | 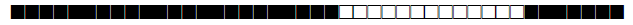 |
777777760000771 | 2 |
| 2047 | LAM | 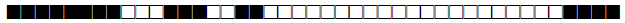 |
776163000000071 | 1 |
| 2329 | Haarlem | 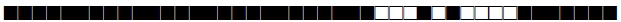 |
777777776120771 | 6 |
| 2330 | LAM10-CAM | 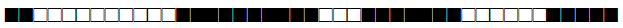 |
600077743760171 | 2 |
| 2331 | LAM | 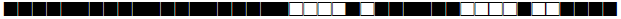 |
777777605760471 | 1 |
| 2332 | T1 | 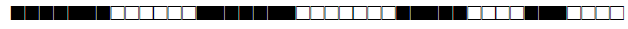 |
774037600760700 | 15 |
| 2333 | Haarlem1 | 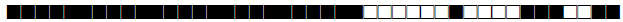 |
777777774020711 | 5 |
| 2336 | LAM | 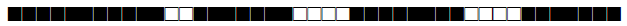 |
777637607760771 | 1 |
| 2337 | Haarlem | 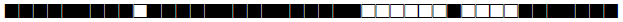 |
777377774020771 | 1 |
| 2338 | Haarlem | 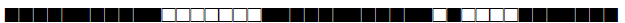 |
777600777720771 | 3 |
| 2341 | LAM | 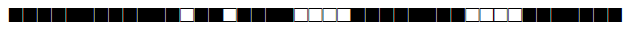 |
777733607760771 | 2 |
| 2372 | LAM1 | 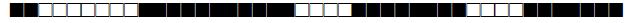 |
600777607760771 | 1 |
| 2516 | S |  |
756377777760771 | 2 |
| 2567 | H1 | 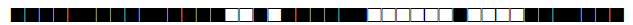 |
777771374020771 | 2 |
| 2576 | LAM | 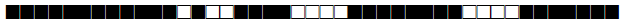 |
777723607760771 | 1 |
| 2647 | LAM | 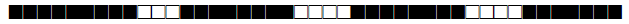 |
777077607760771 | 2 |
| 2836 | UNK | 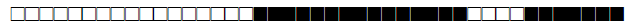 |
000001777760771 | 1 |
| 2891 | UNK | 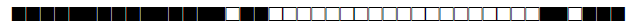 |
777773000000331 | 3 |
| 2892 | Haarlem | 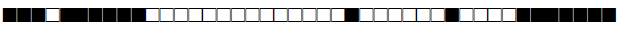 |
737400004020771 | 2 |
| 2893 | LAM10-CAM | 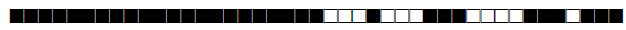 |
777777742160731 | 1 |
| 2894 | LAM | 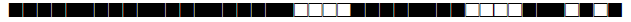 |
777777607760721 | 1 |
| 2895 | Haarlem | 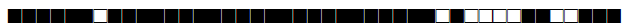 |
773777777720631 | 1 |
| 2896 | LAM | 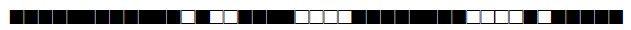 |
777723607760571 | 1 |
| 2897 | LAM | 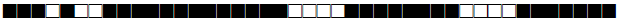 |
723777607760771 | 1 |
| 2898 | T1 | 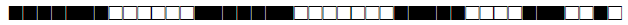 |
774037600760710 | 1 |
| 2899 | T2 | 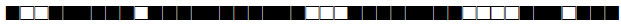 |
477377707760731 | 1 |
| 3051 | UNK | 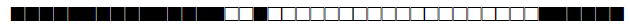 |
400000010020771 | 4 |
| 3072 | UNK | 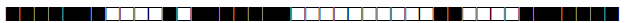 |
776137600020771 | 3 |
| 3073 | T | 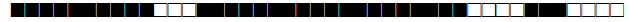 |
777437777760700 | 3 |
| 3074 | UNK | 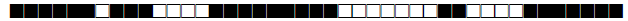 |
773417760060771 | 2 |
| 3075 | LAM | 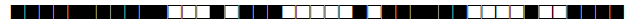 |
777613405660471 | 3 |
| 3076 | T | 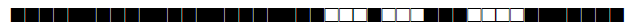 |
777777742160771 | 7 |
| 3077 | H3 | 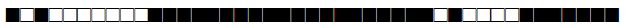 |
500377777720771 | 1 |
| 3078 | H3 | 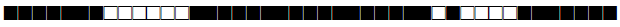 |
774037777720771 | 1 |
Table 1: Description of Shared International Types (SITs) reported for MTBC strains isolated in Morocco from 1997 to 2015.
MTB population structure based on MIRUs-VNTR
Several reports on the incidence and the distribution of tuberculosis in the different communities of Morocco have been published [22], but population genetic studies are scarce. Up to date, four studies using MIRU-VNTR were conducted in Morocco and thus were included in this review. Tazi et al. [23] applied random amplified polymorphic DNA (RAPD) and MIRU-VNTR to analyze the genetic diversity and the population structure of the MTBC isolates circulating in Grand Casablanca. Much more later, the pattern of TB transmission and the risk factors associated with transmission dynamics of this disease were questioned. The study includes 155 isolates of M. tuberculosis (150 patients) mostly collected in 1997 and 1998, with five isolates representing follow-up of five different patients [24].
The results showed an unexpected genetic diversity with a clonal structure for this population in such a high incidence community. The clustering rate was 53.1%, and the recent transmission rate accounted for 37%, taken together, those rates are lower than the ones reported in high incidence TB areas suggesting that reactivation of old TB infections may be responsible for the endemic situation of TB in Casablanca. From a phylogenetic window, there was no association between a sensitive/ drug resistant profile and the clinical characteristics of the disease.
MTB population structure based on both MIRUs-VNTR and spoligotyping
Three national studies provided an overview of the population structure of MTBC in many areas of Morocco and assessed to genetic diversity identified by spoligotyping in combination with MIRUVNTR typing 12 or 24 scheme and its probable association with recent transmission of tuberculosis in Morocco.
Lahlou et al. [14] performed 12-loci MIRU-VNTR typing on 114 strains isolated from patients living in Casablanca. 71 distinct 12- loci MIRU patterns were identified: 48 MITs and 23 orphan patterns; MIT43 (n=15) and MIT 42 (n=7) being the most prevalent. The Hunter and Gaston Discriminatory Index (HGDI) of different MIRU-VNTR loci showed that MIRU10, 23, 26 and 40 were highly discriminant.
Additional subtyping by 12-loci MIRU-VNTR method reduced the clustering rate from 72.8% (by spoligotyping alone) to 29.8% which highlights the potential benefit of the current typing scheme in this setting. Likewise, the recent transmission rate decreased from 64% based on spoligotyping alone to 20.2% suggesting thus that reactivation of latent infection is more common than previously assumed.
Chaoui et al. [14] performed a Subsequent 12-Loci MIRU typing on 185 MTB isolates previously spoligotyped which resulted in a total of 25 SIT/MIT clusters (n=66 isolates, 2-6 isolates per cluster), with a resulting recent transmission rate of 22.3% instead of 65.9% established by Spoligotyping alone. The MIRU-VNTR patterns corresponded to 69 MITs for 138 strains and 46 orphan patterns. The most frequent patterns were MIT43 (n=8), MIT9 (n=7) and MIT42 (n=7). The allelic diversity of individual loci was calculated, only six loci were moderately to highly discriminative namely MIRUs 10; 16; 23; 26 and 40. Hence, MIRUVNTRs 12 loci typing provided a global picture of MTB population structure in Morocco and a preliminary idea about the transmission rate. Although the population structure is quite homogeneous as seen by spoligotyping alone, the number of potential epidemiological links initially overestimated, was considerably reduced as well as a higher level of biodiversity was seen when subclustering by 12 MIRU-VNTRs. Moreover, the calculation of allelic diversity index argues the potential use of spoligotyping in conjunction with optimized 15- or 24-loci MIRU-VNTRs in future investigations to conclude for epi-links [25].
Later on, the population structure of MTBC isolates circulating in Morocco has been evaluated by applying subsequent 24-Locus MIRUVNTR typing on 75 MTB isolates, the latter identified 64 unique types and 11 isolates in 5 clusters (2-3 isolates per cluster), and largely reduced the clustering defined by spoligotyping alone or even by spoligotyping combined with 12-locus MIRU-VNTR typing, from 76% and 48% to 14.6%, respectively.
Therefore, the results argue the use of 24-locus MIRU-VNTR typing in Morocco, to reduce the degree of overestimation of epidemiological links among isolates analyzed by spoligotyping alone or in combination with 12-locus MIRU-VNTR typing.
24-locus MIRU-VNTR typing as well as 12-locus MIRU-VNTR, but at lower degree, are likely beneficial to distinguish MTBC strains sharing highly predominant prototypic spoligotypes namely SIT42, 50 and 53. Of note, not all 24 loci are necessarily required since the calculation of allelic polymorphism index has shown that for 8 of 24 loci had a poor value and could be excluded from future investigations [15].
Based on findings reported in this review and from literature, it would be judicious to carefully evaluate different sets of variable number of tandem repeats loci for genotyping MTBC in Morocco since the number of MIRUs-VNTR loci strongly affects the calculation of recent transmission rate and then to develop routine molecular epidemiological surveillance with a newly discriminatory subset of MIRU-VNTR loci [26,27]. An optimized 15-loci MIRU-VNTR set (but including MIRU23) will be beneficial for epidemiological purposes whereas 24-locus MIRU-VNTR set should be dedicated to phylogenetic studies.
Finally, no relationship between clustering with previous TB treatment/ history or drug resistance exists, implying that most patients develop drug resistant TB individually through selective pressure imposed by inadequately supervised treatment regimens (acquired resistance) rather than through transmission of drug resistant strains (primary resistance) [28,29].
Tuberculosis and populations’ dynamics
Population dynamics, although very common, is a complex phenomenon which influences deeply the global epidemiology of diseases. A closer look to data on Morocco demographics reveals three landmark events (i) Human exchange: massive population outflow from Morocco to European countries since the 1960s and minor migration flow from southern European countries to Morocco; (ii) A mass installation of workers in the northern of Morocco particularly Tangier which is a hotspot areas of TB within the framework of industrialization since the 2000s; (iii) Population outflow from African countries mainly sub-Saharan countries, a process that started in the 2000s which increases the probability of pathogen exchange. Given the intermingling of population and the lack of demographical and epidemiological data for TB in migrants, there is an urgent need to complete TB status especially for undocumented migrants [30].
Molecular fingerprinting of MTBC isolates was recognized as a powerful tool that permits detection of intra transcontinental spread of TB and outbreaks [31]. Within this context, national studies have described a few spoligotypes of particular interest with a biogeographic specificity isolated from young people. Indeed, regarding the historical and geographical proximity between Morocco and Spain, of particular interest was the occurrence of T5-Madrid2 (SIT58 and SIT1227) and LAM12-Madrid1 (SIT209) sub-lineages. Previously found to be characteristic of Spanish-population and Spain related settings [32], their presence in our MTB population reflect migratory movements of people between Spain and Morocco.
Other spoligotypes belonging to the LAM10-CAM lineage, namely prototype SIT61 and its hypothetic variants: SIT2330 and SIT2893 were isolated in Morocco with phylogeographical specificity to Cameroon and neighbouring countries in West Africa. The presence of these genotypes in Morocco is likely attributed to importation by sub- Saharan migrants to and across Morocco [33-36]. This hypothesis is to be confirmed.
In general, it is recognized that immigration affects TB rates in a given country. Indeed, the Country of birth/origin is the best indicator of the risk of developing TB, which is further linked to the degree of endemicity of TB in the migrant’s home country. Moreover, TB rates in a given country are strongly affected by increased number of migrants from highly endemic countries harbouring a large proportion of TB cases [37].
Molecular markers and TB transmission
Molecular markers mainly MIRU-VNTR have been used extensively as they increase the power of discrimination and identify the real proportions of groupings associated with active transmission while recognizing the benefits of the knowledge of the circulating genotypes by spoligotyping. This information is of critical importance to National Tuberculosis Control Program to intensify its control strategies to avoid disease transmission. Moreover, the choice of the molecular marker is crucial with respect to the objectives of a given study. On the other hand, molecular clocks of the markers currently in use should be studied in relation to the predominant lineages observed (Spoligo and MIRU-defined clades); which may be helpful in allocating different weights to each of the markers in a given setting [38].
This review represents the first global snapshot of MTBC genetic diversity from Morocco. Overall, MTBC population structure in Morocco is almost clonal, highly homogeneous and stable. However, the paucity of studies performed of MTBC strains from Morocco implies that additional studies are needed to assign the appropriate weight to each method/marker and to ultimately offer a common/ personalized strategy for future epidemiological investigations. Importantly, a prospective genotyping program has to be set up by National TB program to better evaluate the routes of infection and transmission of TB and to closely monitor MTBC strains as they move is space and time.