Immunome Research
Open Access
ISSN: 1745-7580
ISSN: 1745-7580
Review Article - (2019)Volume 15, Issue 1
The progression in science gives novel and deeper understandings of human beings organisms. The Human Genome Project uncovered about 20,500 human genes. More recently, non-coding RNAs saw the light and gained researchers interest. Among the different subsets of these non-coding RNAs, microRNAs were identified as 18-25 nucleotides long and have been shown to play critical regulatory roles in a wide range of cellular processes. Emerging studies highlighted the importance of miRNAs in health and in disease. Ten years ago, the role of miRNAs in cutaneous system has been established from skin formation in early life to skin homeostasis maintenance. In addition, a deregulated miRNAs profile was shown to cause major skin disorders. Herein, in this review, a global discussion and findings of the different aspects of miRNAs biology will be covered with a focus on the role of miRNAs in skin biology.
The genome and RNA world
Eukaryotic multicellular organisms, such as human beings, owe their existence to the famous double helix of deoxyribonucleic acid (DNA), which codes all of our genes known as our genome. In 2001, the Human Genome Project (HGP) uncovered about 20,500 human genes [1]. This genome dictates our genetic identity our physical appearance (shape, sex, skin color, eye color, etc.) and impacts our physiological behavior (health and disease). The inherent information encoded in the DNA is transcribed into messenger RNA (mRNA) that is subsequently translated into a protein, a process known as the central dogma [2].
Since our cells come from the same zygote, they share the same genes. This begs the following question: what is driving a keratinocyte differentiation from a nerve or a muscle cell? The answer lies in genetic regulations that control the amount of gene expression via an on-off system and any perturbation or deregulation at the genomic level up, has an impact at the protein level resulting sometimes in disease manifestations. This complex regulation process depends on a particular class of noncoding RNA that doesn’t translate into proteins, but instead, functions as the genome’ regulator [3,4].
These non-coding RNAs are divided into two categories, the long and the small non-coding RNAs (sncRNA). They both play a distinctive role whether in RNA editing, chromatin modification or RNA silencing [5-7]. In this review, we focused on a subset of the sncRNAs, the microRNAs (miRNA or miRs). This recently discovered subclass of small ubiquitous molecules, 18-25 nucleotides long, have been shown to play critical regulatory roles in a wide range of cellular processes (Figure 1) [8,9].
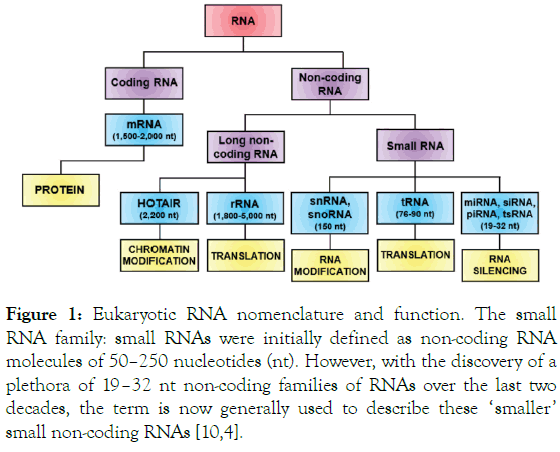
Figure 1. Eukaryotic RNA nomenclature and function. The small RNA family: small RNAs were initially defined as non-coding RNA molecules of 50–250 nucleotides (nt). However, with the discovery of a plethora of 19–32 nt non-coding families of RNAs over the last two decades, the term is now generally used to describe these ‘smaller’ small non-coding RNAs [10,4].
The history of microRNAs
The fundamental understanding of miRNA gene regulation stems from the first miRNA discovered in 1993 in the nematode Caenorhabditis elegans, ‘lin-4’ [11]. C. elegans larval development involves multiple regulatory genes. These genes are responsible for the spatial and temporal configuration of the many cell types generated during the nematode life cycle. Among these genes, lin-4 and lin-14 are known for their central role in the temporal regulation of the cell lineage. Lin-4 is expressed at the end of Larval stage 1 (L1) whereas lin-14 is expressed at throughout the L1 and absent in later stages. Previous studies reported that the normal down-regulation in 1in-14 protein levels during C. elegans development depends on sequences in the 3'-untranslated region (UTR) of the lin-14 mRNA, though the underlying mechanism of regulation was not uncovered at that time [12,13]. Then, Ambros and colleagues, Rosalind Lee and Rhonda Feinbaum, discovered that lin-4 yields a pair of small RNAs of 22 and 61 nucleotides (nt) that fold into a stem loop forming a precursor for a shorter RNA instead of coding for a protein [11]. Moreover, they showed that lin-4 has the ability to regulate lin-14 by binding to multiple sites in the 3’UTR, exerting by that an antisense effect [14]. Consequently, the lin-14 mRNA translation is blocked through the lin-4 complementary binding. Hence, Reducing the lin-14 protein level but not the mRNA resulting in the switch between the stages of larval development (Figure 2) [15].
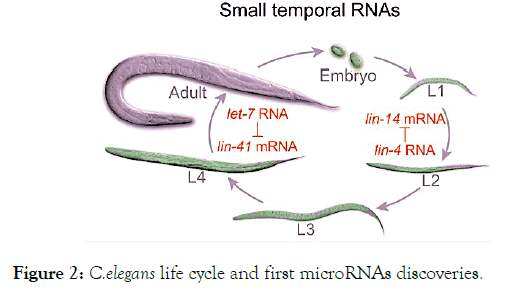
Figure 2. C.elegans life cycle and first microRNAs discoveries.
Seven years later, the second miRNA ‘let-7’ saw the light in C. elegans developmental events going for the cell divisions of L4 to the adult phase by downregulating lin-41 (Figure 2) [16,17]. Only twenty-years after the discovery of lin-4, miRNAs superfamily was recognized as a small non-coding RNA endowed with a highly important role in gene regulation [18-20]. To date, over than 2,500 miRNAs are identified in humans through advances in sequencing technologies and large-scale analysis [21]. According to Lewis D and colleagues, miRNAs target over one third of human genome. In other words it regulates more than 30% of our gene sets. Herein termed the genes fine tuners [22,23]. This elucidates the implication of miRNA in a wide range of physiologic and pathologic events. Hence, understanding miRNAs functioning may be useful for diagnostic and therapeutic applications on corresponding pathologies.
MicroRNAs biogenesis
Canonical biogenesis: In mammalian cells, miRNAs biogenesis is a complex multi-step compartmentalized process in which the transcription starts with Drosha in the nucleus and Dicer in the cytoplasm where maturation occurs [24,25]. The canonical biogenesis of miRNAs can be divided into 5 steps as illustrated (Figure 3).
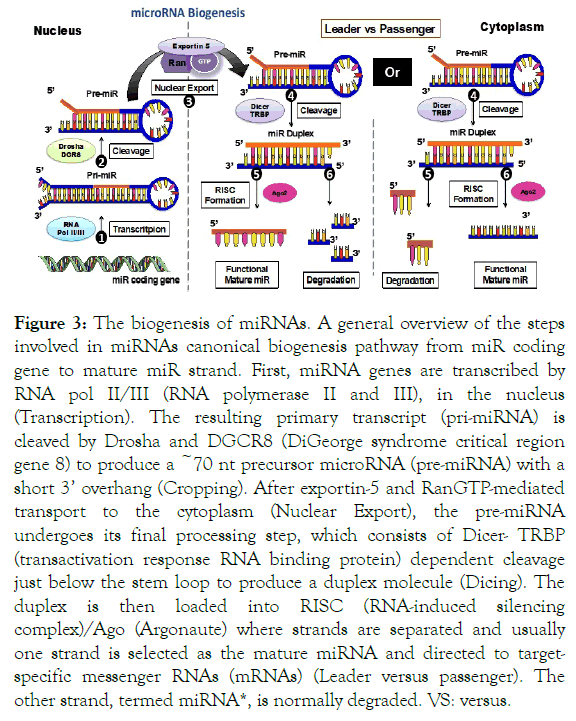
Figure 3. The biogenesis of miRNAs. A general overview of the steps involved in miRNAs canonical biogenesis pathway from miR coding gene to mature miR strand. First, miRNA genes are transcribed by RNA pol II/III (RNA polymerase II and III), in the nucleus (Transcription). The resulting primary transcript (pri-miRNA) is cleaved by Drosha and DGCR8 (DiGeorge syndrome critical region gene 8) to produce a ~70 nt precursor microRNA (pre-miRNA) with a short 3’ overhang (Cropping). After exportin-5 and RanGTP-mediated transport to the cytoplasm (Nuclear Export), the pre-miRNA undergoes its final processing step, which consists of Dicer- TRBP (transactivation response RNA binding protein) dependent cleavage just below the stem loop to produce a duplex molecule (Dicing). The duplex is then loaded into RISC (RNA-induced silencing complex)/Ago (Argonaute) where strands are separated and usually one strand is selected as the mature miRNA and directed to targetspecific messenger RNAs (mRNAs) (Leader versus passenger). The other strand, termed miRNA*, is normally degraded. VS: versus.
Step 1 Transcription: miRNAs loci are found in various genetic contexts originating from different transcription units (TUs). They can be divided into three groups according to their genomic locations and their relative disposition within exons or introns [26]. This suggests slightly different mechanisms of biogenesis depending on the miRNA coding gene origin. The majority of mammalian miRNAs are located in the intronic regions of either non-coding or protein coding TUs. In this case, the same transcript can serve to produce both protein and miRNA. However, exonic miRNA arises only from non-coding TUs. Some miRNAs are transcribed autonomously in which one hairpin is generated from a single miRNA gene [19,18,27]. Others are generated from a polycistronic unit in which a single transcript harbors a cluster of different miRNAs genes that are in close proximity [28-30].
The transcription begins in the nucleus where RNA polymerase II transcribes most of miRNA coding genes (few miRNAs are transcribed by RNA polymerase III) into a large 5’ capped and 3’ polyadenylated primary miRNA transcript (pri-miRNA) [29,30]. The latter is characterized by its stem hairpin loop structure with long sequences extending from the 5’ and 3’ ends.
Step 2 Gatekeeper of the canonical pathway: processing by microprocessor. Before the primary transcript reaches the cytoplasm, it must undergo processing to produce a shorter stem loop structure referred to as precursor-miRNA (pre-miRNA). This occurs via a trimeric complex formation, known as a microprocessor, which is able to recognize the frequently occurring motifs of pri-miRNA (U6 motif at basal segment and UGU/GUG motif at apical loop), and to excise the upper part of the RNA hairpin. The cleavage is mediated by RNase III enzyme Drosha that is stabilized by its binding partner DiGeorge syndrome critical region gene 8 (DGCR8) (also known as Pasha in nematodes) to facilitate processing [31]. Drosha interacts with the basal side of the stem through U6 motif recognition whereas DGCR8 interacts with the upper stem and the apical loop via UGU motif recognition [32-34]. This trimeric complex, composed of Drosha and two DGCR8 proteins, forms the microprocessor system yielding the premiRNA, which is constituted of about 60 to 70 nt.
Step 3 Nuclear export: At this key step, the pre-miRNA associates with exportin-5 complex which is bound to the GTP form of its cofactor Ran [35-38]. The recognition of pre-miRNA by exportin-5 happens through specific structural features that require a 22 base pair (bp) long stem and a short 3’ overhang [39]. The complex subsequently translocates to the cytoplasm across nuclear pore complexes embedded in the nuclear plasma membrane [40]. Once in the cytoplasm, GTP hydrolysis to GDP results in pre-miRNA discharge.
Step 4 Processing to double-strand miRNA generation: The pre-miRNA encounters Dicer, another RNAse III enzyme which lops off the loop yielding around 22 nt miRNA duplex [41]. Dicer uses its Piwi Argonaute Zwille domain (PAZ domain) to interact with the 3 ’ end of pre-miRNA and is stabilized by association with several other proteins such as the Argonaute family [42-44]. The choice of the cleavage site is oriented by a Dicer partner protein also known as transactivation response RNA binding protein (TRBP) [45,46]. This cropping process gives rise to miRNA duplex.
Step 5 miRNA maturation: At this stage, the miRNA duplex is composed of two complementary strands. With the help of the Heat Shock Protein (HSP70/90) chaperone machinery. The conformation Argonaute protein (Ago) is modified to enable miRNA duplex loading into it [47]. Subsequently, the Nterminal domain of Ago induces unwinding of the duplex and only one strand remains to form an effector complex, the RNAinduced silencing complex (RISC) [48]. The strand selection is usually believed to be dependent on the relative thermodynamic stability of the two ends of the duplex. For instance, the low base pairing stability at the 5’ end, favored by mismatch and G, U pairing, promotes the entry of the guide strand into the RISC. There, it exhibits its function of gene regulation [49,50]. Until this day, there exist no clear data regarding the fate and the mechanism of removal of the other unselected complementary or passenger strand (notified by a star (*)). Some studies reported that this passenger strand might be destined either to be degraded as merely carrier strand or to be expressed abundantly as potential functional guide miRNA [51,52]. Once loaded into RISC, miRNAs pair with mRNAs to direct posttranscriptional repression.
Non-canonical biogenesis
The following canonical pathway illustrates the vast majority of miRNA biogenesis process, particularly those that are conserved and abundant. This pathway can be partially used by unconventional miRNA to generate alternative biogenesis pathways classified as non-canonical biogenesis. An example would be the Drosophila short RNA duplexes derived from short intronic hairpins termed "mirtrons" that mimick the structural features of pre-miRNAs. The Mirtrons then bypass Drosha process and merge with the canonical miRNA pathway during hairpin export by exportin-5. After that, they are cleaved by Dicer and incorporated into silencing complexes [53-58].
Regulation of microRNAs occurs from transcription to end stages processing
The miRNA biogenesis pathway is tightly regulated. This is because alterations in the transcriptional and proteomic profiles of the miRNA processing machinery may lead to aberrant expression of specific miRNAs contributing to disease development [59,60]. Therefore, a widespread miRNAs regulation takes place during transcription, Drosha-Dicer processing, and RNA editing. Each step of the biogenesis progression can be considered as a hotspot for regulation [9,61,62].
The transcription of miRNAs can be controlled extensively by Pol II associated factors and epigenetic regulators. This transcription control can either mediates Pol II activity or represses it. This usually occurs to amplify the effect of miRNA in a particular biological context through shared homology between miRNA promoter and transcription factor sequence. For example, p53 transactivates the miR-34 cluster, both known tumor suppressor genes. On the other hand, Myc a tumor promoter gene activates the oncogenic miR-17 cluster and suppresses the tumor suppressor miR-15a cluster [9,61,63].
The regulation at pri-miRNA processing level is achieved on one of 2 sides: either the RNA or the protein side. At the RNA side by interfering with feature element sequences that reside at the basal region and terminal loop region of pri-miRNA (UG motif, CNNC motif and UGUG motif) through RNA Binding The regulation at pri-miRNA processing level is achieved on one of 2 sides: either the RNA or the protein side. At the RNA side by interfering with feature element sequences that reside at the basal region and terminal loop region of pri-miRNA (UG motif, CNNC motif and UGUG motif) through RNA Binding Protein (RBP) and RNA modifying enzymes [64]. For instance, lin-28 interacts specifically with let-7 family and KH-type splicing regulatory protein (KSRP) a splicer factor that interacts with the loop to block pri-miRNA processing [64,65]. Dead box helicase (DDX) proteins seem to interact with the single stranded region leading to increased processing by Drosha [66]. Lastly, the adenosine deaminase acting on RNA enzymes (ADAR1 and ADAR2) mediate RNA stem editing from adenosine to inosine, which interferes with Drosha processing [67-69]. The protein side regulations occur by targeting the microprocessor proteins through phosphorylation, acetylation or ubiquitinilation. All of which are important hallmarks for protein stability, nuclear localization and processing activity of Microprocessor. The nuclear localization of Drosha is ensured by its phosphorylation via glycogen synthase kinase 3β (GSK3β) [70,71]. Similarly, the acetylation of Drosha is essential to inhibit its degradation and to consequently provide stability maintenance [72]. On the other hand, DGCR8 ’ s increased stability is dependent on phosphorylation by extracellular signal-regulated kinases (ERK) and increased affinity to pri-miRNAs which is in turn dependent on deacetylation by histone deacetylase 1 (HDAC1) [73,74].
The pre-miRNAs are regulated in a similar fashion. Pre-miRNAs editing influences the miRNAs stability and abundance. Some pre-miRNAs expression can be controlled by lin-28 binding to pre-miRNAs which induces terminal uridyltransferase activity by Terminal Uridylyl Transferases (TUT4/TUT7) that in turn mediates decay via Dis3 Like Exonuclease 2 (DIS3L2) [75-78]. Likewise, BCDIN3 domain contains RNA methyltransferase (BCDIN3D), a methyltransferase that regulates negatively miRNA maturation via O-methylation of the 5' monophosphate resulting in Dicer pre-miRNA processing blockade [79,80]. Another regulatory mechanism to control miRNAs expression includes endonucleases action. For example, Monocytes chemotactic protein 1 induced protein 1 (MCPIP1) an endonuclease that can cleave the terminal loops of pre-miRNAs to negatively control miRNAs biogenesis [80]. This kind of regulation seems to be important in cancer and immune response [48]. Since TRBP assists Dicer to accurately recognize the cleavage site of pre-miRNAs, it can be considered as major key modulator of the processing efficiency and cleavage sites of some pre-miRNAs to yield variable mature miRNAs isoforms [81,82]. Moreover, TRBP can be phosphorylated by ERK leading to its enhanced stability [83].
The RISC formation is another important step where Ago stability and localization are regulated through hydroxylation by type I Prolyl 4-hydroxylation (4PH), phosphorylation by Mitogen Activated Protein Kinase activated protein kinase 2 (MAPKAPK2), Poly ADP-ribosylation and ubiquitination [84-88]. Furthermore, the most critical regulation happens at the mature miRNAs stability layer. In general, in steady state conditions, miRNAs are very stable with a long half-life. Nonetheless, under some defined biological systems (e.g. cancer, hypoxia, inflammatory stress, development, cell cycle, etc), the miRNAs half-life can be shortened and can present increased turnover. Some RNA-modifying enzymes such as exoribonucleases XRN1/XRN2 in C. elegans are involved in accelerated miRNA turn-over [89,90]. As such, the adenylation modification by Wispy, as well as the methylation by Hua enhancer 1 (Hen1) of the 3’ end of the miRNA can modulate miRNA stability [91-93].
MicroRNAs function as gene fine tuners
miRNAs act by sequence complementarity pairing with mRNA molecules. Two distinct mechanisms are involved depending on the degree of the complementarity between both miRNA sequence and its mRNA target [94]. When the complementarity is perfect (pre-dominant phenomenon in plants), the RISC complex ensuring the interaction between the miRNA and the mRNA allows the action of ribonucleases capable of cleaving, and therefore of degrading, the mRNA molecule (Figure 4) [42,95].
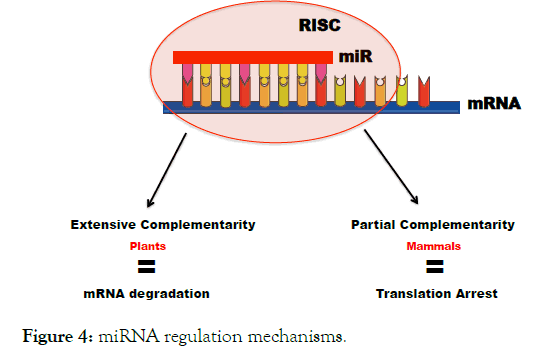
Figure 4. miRNA regulation mechanisms.
It is, in this case, a control at the transcriptional level. When the complementarity is partial (a pre-dominant phenomenon in mammals), the mRNA is not cleaved, But the binding of miRNA to the 3’UTR mRNA causes translation repression. In both cases, the consequence is an extinction of the expression of the corresponding gene as a result of translation arrest or mRNA destabilization or a combination of the two [11,14,96]. The repression mode in humans relies primarily on a main adpator protein called TNRC6 (trinucleotide repeat containing 6) that exists in 3 paralogs A, B and C [97-102]. The TNRC6 recruited by the silencing complex acts via three complementary regulatory mechanisms:
1) Induction of the poly (A) shortening through its interaction with the poly (A) binding protein complex (PABPC). This results in deadenylase activity enhancement responsible of the decapping and 5’ to 3’ exonucleolytic decay resulting in mRNA destabilisation [99,103].
2) Reduction of translation efficiency mediated by Carbon Carbolite Repressor 4 – Negative On TATA (CCR4-NOT) and its DDX6 complex recruitement that causes translation arrest via binding to the decapping complex [99,100,104].
3) Repression of translation and enhancement of decay via DDX6 interference with the translation machinery. This happens through a competition with eukaryotic translation initiation factor 4 gamma (eIF4G) for eukaryotic translation initiation factor 4E (eIF4E) binding caused by DDX6 interaction with eIF4E transporter [105-108].
The ability for a miRNA molecule to bind mRNA in imperfect sequence complementarity allows a multitude of possible targets. Some algorithms derived from bioinformatics predict up to 200 genes whose expression would be potentially controlled by a given miRNA molecule. Thus allowing a third of the genome to be regulated by these small non-coding RNA molecules, which underlines the importance of this control mechanism for the cell [8,109,110]
miRNA target recognition
Initial understandings for miRNA - mRNA target recognition are derived from studying the first miRNAs lin-4 and let-7. The reported data back then suggested the presence of multiple conserved sites within the 3’UTR of mRNA coding for lin-14 and lin-41, where lin-4 and let-7 respectively have sequence complementarity [11,13,14,16,17,19]. Nowadays, the target sites of miRNAs are separated into three. The canonical, atypical and non-canonical sites (Figure 5) [8].
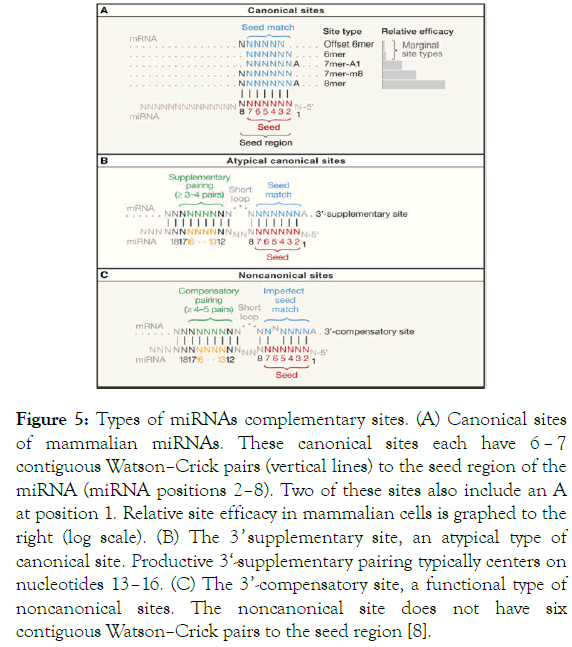
Figure 5. Types of miRNAs complementary sites. (A) Canonical sites of mammalian miRNAs. These canonical sites each have 6 – 7 contiguous Watson–Crick pairs (vertical lines) to the seed region of the miRNA (miRNA positions 2–8). Two of these sites also include an A at position 1. Relative site efficacy in mammalian cells is graphed to the right (log scale). (B) The 3 ’ supplementary site, an atypical type of canonical site. Productive 3‘-supplementary pairing typically centers on nucleotides 13–16. (C) The 3’-compensatory site, a functional type of noncanonical sites. The noncanonical site does not have six contiguous Watson–Crick pairs to the seed region [8].
The core region in which the interaction between miRNA and mRNA happens is called the ‘seed region’. There, the pairing is attained at the most conserved region, the 5’ end of miRNAs centered between nt 2 to 8 [8,110]. The recognition within this canonical site is associated with 5 site types including 8mer, 7mer-m8, 7mer-A1, 6mer, and offset 6mer. In this canonical site, the pairing can occur according to Watson-Crick pairing between nt 2 to 7 leading to 6 sites match (6mer) or through an offset pairing by one nt (offset 6mer). Also, these regulatory sites can have either an additional match to miRNA nt 8 (7mer – m8) or an A pairing with nt 1 (7mer-A1), or both (8mer), to make 7 or 8 nt matching sites, respectively [109,111,112]. The best repression goes to the highest efficient sites with perfect matching that include the 7 and 8 sites-match. Whereas the least effective goes to the 6 match sites [111,113-115].
Around 5% of this seed match sites can have a supplemental pairing at 3 ’ region of the miRNA involving nt 13 to 16 [110,114]. This form of recognition is classified among the atypical sites. Nonetheless, this additional pairing has little influence on the affinity and efficacy of miRNA-mRNA recognition [8,116,117].
The last type of miRNAs complementary sites is the noncanonical site. It accounts for less than 1% of the preferentially conserved miRNA sites in mammalian miRNA. Complementarity at this site happens when the seed core doesn ’ t fulfill the Watson-Crick matching. Instead, a compensatory 3’ pairing is set typically across nt 13 to 16 to counterbalance the missed 6 contiguous pairing that causes the imperfect match [8,110,114]. To date, no direct scientific proof explains this type of recognition. This rare 3’ pairing could be considered as highly secured regulation system when one mRNA requires regulation by a family of miRNAs. To avoid competition between miRNAs, one particular miRNA can be programmed to sit on the seed region and another miRNA to sit on 3’ region affording a double regulation system.
miRNA target prediction strategies
Several difficulties arise from miRNA studies rendering prediction of miRNAs regulatory targets delicate [110]. First, one miRNA can target several mRNA. Second, the evolution of next generation sequencing (NGS) technologies generated massive amounts of data. Third, the function of miRNA can vary depending on the organ and the physiological condition [114]. Fourth, other parameters including RNA integrity and experimental conditions may influence miRNA expression [118]. Fifth, an expressed miRNA doesn’t prove its functionality; hence, it requires additional characterization studies [119]. Over the years, several bioinformatics analysis programs have been developed to interrogate and interpret deep sequencing data. In 2002, a common public database “miRBase” was created. This unified the definition and nomenclature of miRNAs [21,120-122] and provided access to sequences which formed a library providing information on the already identified miRNAs. Of these, the TargetScan (http://www.targetscan.org) [109,123], PicTar (http://pictar.mdc-berlin.de) [112], miRWalk (http://mirwalk.uni-hd.de) [124] and RNAhybrid (http:// bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html) [125], ComiR (http://www.benoslab.pitt.edu/comir) [126], etc. constitute highly sophisticated systems that continue to develop. With time, the extent and significance of miRNA regulation of gene expression is expected to evolve and become even more evident. Multiple features come in hand to reliably predict targets of a miRNA and to avoid false positives. A contemplated approach would be to follow the broader to narrower funnel type filter: from canonical Watson-crick 3’UTR match [110] to evolutionary conserved 3 ’ UTR sites, consider type, position, number of sites and potential 3’ supplementary pairing [127], and finally crosslinking results from the different bioinformatics tools is essential. This methodology takes into consideration determinants beyond seed pairing. Multiple studies reported the importance of considering structurally accessible sites for pairing, proximity to sites for coexpressed miRNAs and, positioning of the 3’UTR with respect to stop codon and the presence of additional marginal sites in 3’UTR [8,110,127-129]. There are plenty of possibilities, yet the target prediction methods and strategies have long road to improvement.
miRNAs diverse functions in diseases
As miRNAs biology gained the interests of researchers, in 2002, Lagos-Quintana and colleagues showed a tissue specific miRNA signature pointing off a potential connection between miRNAs and diseases [130]. In 2003, the importance of miRNAs development was demonstrated in zebra fish that failed to develop when Dicer-1 gene was inactivated [131]. Then, dicer knocked-out (KO) mice displayed embryonic lethality before gastrulation [132]. Also, Drosophila mutants lacking Dicer-1 in the germline stem cells highlighted the significance of miRNA pathway to bypass Gap 1/Synthesis (G1/S) checkpoint resulting in cell cycle progression and stem cell division [132]. Besides the widespread roles of miRNAs in processes like cell fate determination, axis formation, and cell differentiation for zebra fish development, they were shown to be required for brain morphogenesis in zebra fish embryos [133]. Afterwards, microarray analysis showed that miRNAs downregulate a large number of transcripts population to define metazoan cell types [134]. This result reinforced the previously reported functions of miRNA to prevent cell division and drive terminal differentiation in C. elegans development [11,14,16,17]. In brief, these studies indicated that Dicer and subsequently miRNAs are essential for morphogenesis and vertebrates development.
Moving forward, studies gradually shifted towards functional characterization of miRNAs in human diseases. Profiling studies demonstrated a causal role of deregulated miRNAs profile in the generation or maintenance of tumors [135]. Cancer cells present differential upregulated or downregulated miRNAs signature compared to normal cells, suggesting their role as oncogenic or tumor-suppressor genes. The first evidence of miRNAs regulation in cancer was shown in B cell chronic lymphocytic leukemia (CLL) where a frequent deletion of chromosome 13 containing genes for miR-15 and miR-16 resulted in their function reduction [136]. Further studies demonstrated the correlation between aberrant miRNA expression patterns and increased occurrence of different types of cancers notably miR-125, miR-145, miR-21, miR-155, etc. [137-143]. Cancer growth has been the most prominent example of human diseases with a clear role for miRNA regulation. Thus, miRNAs in malignancies are now considered and used as biomarkers for early diagnosis as well as in the development of novel anti-cancer strategies.
It is only few years ago that efforts to explore the role of miRNAs in non-cancerous pathologies such as metabolic, cardiovascular, neurodegenerative, infectious, inflammatory and autoimmune diseases have been employed. In metabolic disorders, a dynamic bidirectional regulation between miRNAs and glucose metabolism has been uncovered through miR-375, which suppresses insulin secretion [144]. Also, a large panel of miRNAs including miR-143, miR-130a, miR-27, miR-34, miR-224 were shown to participate in adipocyte differentiation, to be involved in lipid and fat metabolism thus, associated with metabolic disorders such as diabetes and morbid obesity [145-150]. In cardiovascular diseases, contribution of miRNAs in particular miR-1 and miR-206 have been identified to have a crucial role in normal cardiac cell function [151]. Moreover, several miRNAs including miR-133, miR-548, miR-320 and miR-155 were found to modulate cardiomyocytes’ activity and vasculature contributing to pathologies such as cardiomyopathy and atherosclerosis [152-157]. miRNAs are fundamental for appropriate brain development [133]. Deregulation in the miRNAs expression pattern can result in major neurodegenerative diseases. Namely, Alzheimer disease (AD), Parkinson disease (PD) and Huntington disease (HD) [158]. In AD, miR-15, miR-146, miR-107, miR-29, miR-106, and miR-9 act by favoring plaques deposition via targeting secretases and neurofibrillary tangles formation thus promoting Tau proteins phosphorylation [159-163]. In PD, brain miRNAs (miR-133 and miR-205) and circulating miRNAs (miR-22-3p, miR-29a, miR-16-2-3p, miR-26a-3p and miR-30) have been associated directly or indirectly with the regulation of α-synuclein or dopamine transport [164-167]. Interestingly, in HD the mutant huntingtin gene that causes neurotoxic effect on a molecular level, inhibits the RISC formation during biogenesis through interaction with Ago leading to reduced miRNAs effector function [168]. On the other hand, the circulating miR-34b was shown to be upregulated in HD patients and seems to exert a protective action by blocking the mutant huntingtin in vitro [169]. miRNAs are emerging regulators of innate and adaptive immunity [170]. During bacterial or viral pathogen invasion, panels of miRNAs are produced comprising mainly of miR-146 and miR-155 that regulate the immune response by impacting cytokines signaling pathway [171,172]. Some studies also reported involvement of miRNAs in the development and differentiation of immune cells such as T and B lymphocytes [172-174]. Furthermore, Growing evidence is suggesting the association of a deregulated miRnome with autoimmune inflammatory diseases (AID) like psoriasis (refer to section 7), rheumatoid arthritis (RA), multiple sclerosis (MS), systemic lupus erythematosus (SLE), inflammatory bowel disease (IBD), etc. [175,176]. In RA, miRNAs interfere at different stages of the pathogenesis. For example, miR-34-5p and miR-34-3p regulate synovial fibroblast whereas miR-124, miR-346, and miR-203 modulate synovial inflammatory niche [177-180]. In MS, miR-326 was shown to play an important role in TH17 polarization; and differential expression of miRNAs (miR-18b, miR-599, and miR-493) was noted to be correlated with MS relapses [181,182]. Microarray analysis of miRNA expression in peripheral blood cells of SLE patients revealed that 16 miRNAs were differentially expressed and miR-146, miR-21 and miR-126 are the most relevant [183]. miR-146a represses the downstream transactivation of type I IFN at the molecular level and targets IFN regulatory factor 5 and STAT-1 while miR-21 and miR-126 regulate DNA methylation leading to SLE [184-186]. In Inflammatory bowel diseases (IBD), a complex interplay between several miRNAs that regulate a network of genes was found to be implicated in inflammatory response (miR-4284, miR-126, miR-29, miR-15, miR-106, miR-10, etc.), in barrier function (miR-21, miR-150, miR-200b) and autophagy (miR-30c, miR-181a, miR-93, miR-106b, etc.) [187-190].
Further ongoing comprehensive studies are being conducted to understand and dissect the complex nature of miRNAs regulatory interactions with the multipath regulatory network in diseases. miRNAs specifically occupy a precious role in skin biology, since the skin is a highly and tightly regulated dynamic organ that consists of a constant cell renewal, regeneration and immune response to maintain cutaneous homeostasis and barrier function. In the following paragraph, an overview of miRNAs ‘interplay in skin biology’ will be presented.
miRNAs and skin
The understanding of miRNA-mediated control in the skin relies on functional studies targeting critical components of the miRNAs ’ biogenesis pathway and on exploring the role of individual miRNAs.
The lethality of constitutive dicer deletion drove the development of Dicer conditional KO (cKO) mice in the skin using the Cre-Lox system [191]. a k14-Cre cKO mouse of Dicer was engineered; making henceforth the studying the role of dicer depletion along embryonic and adult epidermal development and hair follicle (HF) morphogenesis possible. k14- Cre cKO of Dicer displayed abnormal HF expansion, enriched apoptosis in hair bulbs, rough dehydrated skin and neonatal lethality [192]. Although, it was not yet clear which signaling pathways were altered in these cKO mice, it was obvious that Dicer played a critical role in skin development and the morphogenesis of HFs [192,193]. Besides these observations, Dicer/k14-Cre cKO presented hyperproliferative phenotype in the epidermis suggesting the implication of miRNAs in proliferation control and cell cycle regulation during epidermal differentiation [193,194]. Furthermore, k14-Cre cKO mice of Dgcr8 was generated and showed similar phenotype to k14-Cre cKO mice of Dicer as well as comparable miRNA expression patterns [195]. With these findings, the global importance of miRNAs in skin development was revealed and their role in morphogenesis regulation, maintenance of HF stemness, epidermal proliferation and apoptosis was evident. So, researchers ’ interest moved to the functions of individual miRNAs in the skin.
The role of miRNAs in regulating the varied functions of the skin is not yet fully characterized and work is in progress to elucidate the related mechanisms. In the following paragraphs, we/I discuss a general overview of the diverse miRNAs regulatory activity in different context of cutaneous behavior.
Ten years ago, miR-203 was the first miRNA investigated in the skin because of its spatio-temporal expression [194]. Throughout epidermal development, as KCs move forward from the basal layer and become differentiating cells, miR-203 expression intensifies. Highlighting by that a potential role of miR-203 in controlling cellular differentiation and proliferation balance. Indeed, miR-203 targets p63 an essential regulator of stem cell maintenance in stratified tissues resulting in its down regulation and consequently in proliferation inhibition [194,196,197]. Hence, miR-203 acts as a switch between proliferation and differentiation [194]. Accordingly, miR-203 can be considered as a tumor suppressor since its induction in squamous-cell carcinoma cell line resulted in cell proliferation inhibition [197-201]. In psoriasis, which is a hyperproliferative disease, miR-203 expression was augmented [202]. In this particular case, the increased production of miR-203 can be considered as a defense mechanism established to counterbalance the hyperproliferative activity in KCs, yet further studies are needed to comprehend these observations.
The major role of the skin is to shield the organism from external environment and to prevent water loss by exerting barrier functions. The integumentary system is made up of a network of tight junction and structural protein forming a wellstructured envelop leading to epidermal resistance [203,204]. Among these protein networks, E-cadherin (an adhesion molecule) plays a determinant role in skin architecture by ensuring proper localization maintenance of tight junction network [204-206]. Studies have reported the tight control of Ecadherin expression by miR-200 and miR-205 miRNAs through targeting the mRNAs of E-cadherin transcriptional repressors ZEB1 and ZEB2. Thereby, this positive E-cadherin regulation seems to be essential in maintaining epithelial stability [207,208].
Protection against UV radiation happens through pigmentation. This process involves production and dispersion of melanin by epidermal melanocytes to neighboring KCs and vitamin D regulation [209,210]. miRNAs regulate melanogenesis by targeting microphthalmia transcription factor (MITF), a key regulator of color genes coding for Tyrosinase (Tyr) and Hyalurodinase (Hyal). While miR-25 suppresses MITF, miR-21-5p regulates MITF indirectly via Sox-5 and miR-434-5p targets Tyr and Hal to control melanin production [211-213].
Wound healing for instance, is a multistep process that implicates various important cellular and molecular actors. It involves three complementary phases [214-217].
1) Inflammatory phase, which begins with leukocytes, mainly neutrophils in addition to other immune cells, extravasation into wound through damaged blood vessel. Cytokines, chemokines, AMPs and growth factors production then follows to resolve and to clump the lesion. This chemoattraction occurs to achieve angiogenesis [217].
2) Proliferative phase: The aim of this stage is to re-epithelialize the damaged area. It occurs via keratinocytes migration and proliferation. Growth factors and other mediators stimulate the re-epithelization process [218].
3) Remodeling phase: At this point, the skin completes wounding by restoring its architecture and elasticity. Thus, this phase requires an appropriate production and deposition of extracellular matrix (ECM) proteins in particular collagen [217].
The miRNAs microarray profiling identified a deregulated expression of miRNAs during wounding (219). Thereafter, miRNAs were identified as important players in each phase of wound healing process. First, miRNAs can regulate and be regulated by inflammatory mediators during inflammation [219-222]. For instance, after wounding, miR-31 expression is induced in KCs by TGF-β or TNF-α and IL-6 via STAT-3 and NF-κB signaling. miR-31 upregulates Ras/MAPK pathway activity by targeting its negative regulators (Rasa1, Spred1/2 and Spry4) to promote cellular proliferation and migration [223]. miR-31 silencing epithelial membrane protein 1 (EMP-1) is another way to regulate KC proliferation and migration during wound healing [224]. Moreover, miR-21 inhibition was found to cause significant delay of wound closure and skin reconstruction [219]. miR-21 induced by IL-6 promotes cellular migration and proliferation by targeting PTEN and PDCD4 [143,225,226]. Besides, other miRNAs interfere with the inflammatory response such as miR-146 induced by TNF-α and IL-1β targets IRAK and COX2 while miR-155 silences SHIP1, SOCS1, and IL-12 [227,228]. The process of angiogenesis requires endothelial cells proliferation, migration and development that can be regulated in a specific manner by miRNAs. Some of the most well-characterized miRNA-regulated proteins involved in angiogenesis are Spred1 by miR-126 [229], c-kit by miR-221/222 [230], Tsp-1 by miR-17-92 [231,232], ITGB5 by miR-92a [233,234], VEGF by miR-20a [235], and TIMP-1 by miR-17-5p [236]. During re-epithelialization, miR-210 is a major miRNA that profoundly influences KC proliferation that impacts wound closure. miR-210 induced in hypoxic conditions limits proliferation by downregulating cell cycle regulatory protein E2F3 [237]. In remodeling phase, miR-29 plays a key regulation of collagen deposition [114].
In the skin, miRNAs have a versatile range of abilities to interfere with several physiological processes such as KCs proliferation, pigmentation, skin ageing, wound healing, SIS, cutaneous microbiota, etc. Therefore, a deregulation in miRNAs expression can result in skin disorders. miRNAs in skin diseases (Figure 6).
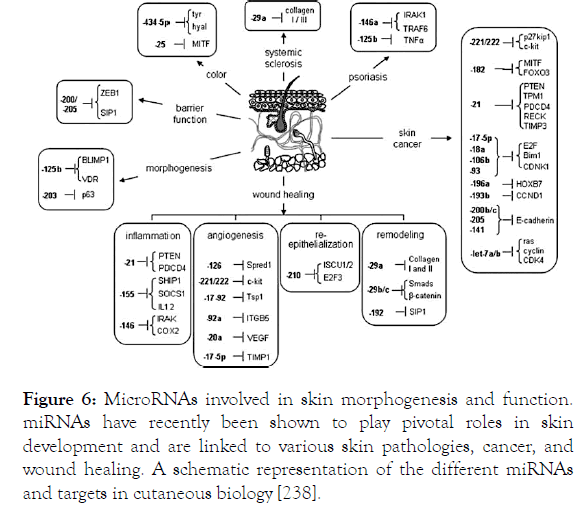
Figure 6. MicroRNAs involved in skin morphogenesis and function. miRNAs have recently been shown to play pivotal roles in skin development and are linked to various skin pathologies, cancer, and wound healing. A schematic representation of the different miRNAs and targets in cutaneous biology [238].
Fibrotic skin diseases
Fibrotic skin diseases is a family of skin disorders including all forms of scleroderma, graft-versus-host disease (GvHD), dermopathy, mixed connective tissue disease, scleromyxedema, scleredema, eosinophilic fasciitis, keloids and hypertrophic scars caused by abnormal wound healing [239,240]. This skin condition is due to cutaneous epithelial hyperproliferation and excessive ECM production. Studies have shown miR-21 as a common overexpressed miRNA between the several fibrotic skin diseases [241,242]. miR-21 regulates at distinctive levels multiple fibrotic diseases because of its pleiotropic potential of miR-21 to control several biological processes in relation with proliferation, differentiation and epithelial-mesenchymal transition (EMT). As a matter of fact, TGF-β the most important profibrotic cytokine, stimulates miR-21 production resulting in hypertrophic scar via targeting PTEN/PI3k/Akt and Smad7 pathway [241,243]. In the same manner, miR-21 induces proliferation and transdifferentiation of keloids fibroblasts and favors EMT of keloids KCs [244-246]. Recent study showed that miR-21 regulates the apoptosis of keloid fibroblasts via targeting FasL and caspase-8 and the mitochondria-mediated apoptoticsignaling pathway [247]. In systemic sclerosis, miR-21 promotes fibroblast proliferation by targeting Bcl2 and Smad7 [248,249].
Skin malignancy
Skin cancer types are divided into melanoma and nonmelanoma cancer. Both types are caused by abnormalities in proliferation, migration and apoptosis of melanocytes for melanoma type and from KCs for non-melanoma form, which can be controlled by miRNAs. Microarray analysis provided a specific signature of miRNAs for cutaneous malignancy [250]. The most relevant miRNAs and miRNAs targets in skin cancer regulation are summarized (Table 1).
| miRNA | Target gene | Expression | References |
|---|---|---|---|
| miR-21 | Bcl2, PTEN, GRHL3 | Up | [251-253] |
| miR-124 | ERK1/2 | Down | [253] |
| miR-214 | |||
| miR-130 | Bcl2 | Up | [254] |
| miR-203 | p63 | Down | [199] |
| miR-210 | PTPN1, TP53I11, HOXA1 | Up | [255-257] |
| miR-29c | DNMT3A, DNMT3B | Down | [254] |
Table 1: List of most commonly implicated miRNAs in skin cancers and their targets.
microRNAs in Psoriasis
In 2007, for the first time, Sonkoly et al. analyzed the miRNA expression profile of psoriasis lesional skin and identified a set of miRNAs to be differentially expressed. Then, these results were further confirmed with other studies from different groups [251-260]. In 2013 and 2014, a differential profile of circulating miRNA from psoriatic patients was shown compared to healthy subjects [261,262]. Since, many of the deregulated miRNAs were studied in functional details in the pathogenesis of psoriasis. In this project, the focus will be on miR-21 and miR-31, which are the most upregulated miRNAs in psoriasis.
One of the first studied miRNAs was the highest upregulated miR-203. In psoriasis, miR-203 targets suppressor of cytokine signaling-3 (SOCS-3) that negatively regulates STAT-3 pathway [202,263]. The upregulation of miR-203 in human keratinocytes (suprabasal layer) is required for their differentiation and is dependent on the activation of the PKC/AP-1 pathway [264,265]. Further studies are needed to reconcile the disparity of miR-203 functions in order to determine its implication and role in the inflammatory and proliferative responses. Xu et al. reported that miR-203 expression was induced by upregulated IL-17. This in turn activates the Janus kinase signaling pathway, which results in VEGF production by immortalized human keratinocytes HaCaT [266].
miR-21 is one of the most abundant miRNAs in psoriasis [202,220,258]. First, miR-21 was demonstrated to suppress to Tcell apoptosis [173]. Then, the Guinea-Viniegra [266] team showed a correlation between miR-21 and the TNF-α pathway. Indeed, activation of the TNF-α signaling pathway is dependent on the availability of the soluble form of the TNF-α cytokine (sTNF) which is ensured by the enzymatic activity of TACE. The latter is blocked by the inhibitor TIMP-3. In an inflammatory context, induction of the TNF-α pathway results in the production of IL-6 which will activate the JNK/STAT pathway causing increased expression of miR-21. This results in a decrease in TIMP-3 and thus in the elimination of TNF-α pathway inhibition. Consequently, uncontrolled activation of TNF-α results in increased psoriasis inflammatory response. Interestingly, targeting of miR-21 in mouse models of psoriasis by miRNA inhibitors could ameliorate the disease phenotype and showed a similar efficacy as anti-TNF therapy [266].
miR-31 is another overexpressed miRNAs in psoriasis that modulates the inflammatory environment. In KCs, TGF-β induced miR-31 targets STK40, a negative regulator of the NF- κB pathway resulting in enriched cytokines and chemokines milieu that contribute in endothelial cell activation, leukocyte attraction and skin inflammation [267]. Another elegant study, reported the protein phosphatase 6 (ppp6c) an inhibitor of the G1-S phase transition in the cell cycle, which is diminished in human psoriatic epidermis, as miR-31 target. miR-31 induced ppp6c suppression results in KCs hyperproliferation [268]. Thus, inhibiting miR-31 maybe a novel therapeutic option to treat psoriasis.
By contrast, other miRNAs are downregulated in psoriasis. They play central roles in regulating the proliferation and differentiation of KCs by mainly acting on growth/transcription factors or cytokines receptors. miR-125 was found to be downregulated both in blood and skin lesions of psoriatic patients. It regulates fibroblast growth factor receptor 2, which suppresses cellular proliferation and prolongs the cellular differentiation of psoriatic KCs [269].The inhibition of miR-125 in human KCs may result in human KCs hyperproliferation and delayed differentiation [270].
In the upper layer of the epidermis of psoriatic skin, miR-99 is another downregulated miRNA. It acts on IGF-1R expression and inhibits keratinocyte proliferation. A decrease of miR-99 leads to an abnormal proliferation of basal layer cells in psoriatic patients [271].
miR-520 is another miRNA that is highly downregulated in psoriatic lesions [272]. It is involved in the regulation of KCs proliferation by inhibiting AKT signaling and downregulating the transcription factor E2F, consequently, stopping the cell cycle progression [273].
Several other miRNAs (e.g. miR-125, miR-99a, miR-424, miR-7, miR-135, etc.) have been identified in psoriasis and shown to modulate the proliferative capacity of KCs [274]. Altogether, these studies underline the weight of miRNAs in psoriasis and their prospective involvement during disease pathogenesis.
Controlling miRNA activity exogenously
When studying the functional role of a miRNA, both gain-offunction and loss-of-function observations are required. To reveal the miRNA’s implication in development, homeostasis and disease, specific tools were developed to modulate their expression whether by upregulation or downregulation [275]. A design of chemically synthesized double-stranded or singlestranded RNA that interferes with the RISC or with the mature miRNA strand either to imitate or to silence endogenous miRNAs was achieved. Consequently, two different classes of synthetic miRNAs also called oligomiR exist: Inhibitors and Mimics (Figure 7).
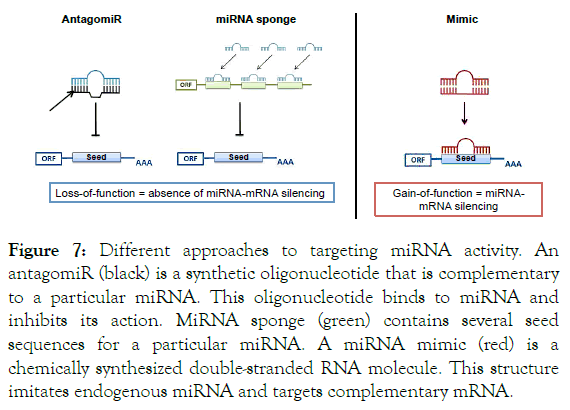
Figure 7. Different approaches to targeting miRNA activity. An antagomiR (black) is a synthetic oligonucleotide that is complementary to a particular miRNA. This oligonucleotide binds to miRNA and inhibits its action. MiRNA sponge (green) contains several seed sequences for a particular miRNA. A miRNA mimic (red) is a chemically synthesized double-stranded RNA molecule. This structure imitates endogenous miRNA and targets complementary mRNA.
miRNA inhibitors are chemically modified, single stranded nucleic acids. They are designed to specifically bind to and inhibit endogenous miRNA molecules. The introduction of miRNA inhibitors into a biological system results in a loss-offunction assay with a predicted decrease in the function of the targeted endogenous miRNA. Two types of inhibitors are available: synthetic and expressed inhibitors [276]. The synthetic inhibitors (referred as antagomiR) are generally nonhydrolyzable, single-strand complementary to the mature miRNA. The inhibition mechanism is mediated by irreversible binding of the inhibitor to the mature miRNA-loaded RISC. Successively, the interaction of the mature miRNA with its endogenous mRNA targets is prevented. Expressed inhibitors, however, known as miRNA sponges, are generally artificial mRNA constructs with multiple miRNA sites. These miRNA sponges function by competing with natural mRNA targets to bind miRNAs (Figure 6) [277,278]. The usage of a combination of both types inhibitors to induce loss-of-function increases the likelihood of detecting associated phenotypes with neutralized miRNA. In some cases, those tools can be used to silence aberrantly expressed miRNAs [275-277,279-282].
miRNA mimics are small, chemically modified double-stranded RNAs that mimic endogenous miRNAs duplex and enable miRNA functional analysis by activity up-regulation. miRNA mimics are used experimentally to supplement endogenous miRNA expression, for gain-of-function studies. Synthetic miRNA mimics on the other hand, are chemically synthesized; double-strand RNA molecules that are intended to mimic the transient duplexed product of Dicer complex processing [283,284]. Like naturally occurring miRNAs, the two strands of a synthetic mimic unwind, and the single-strand mature miRNA is incorporated into RISC to down-regulate mRNA transcripts (Figure 6) [285]. Synthetic mimics can be chemically modified to favor loading of the mature miRNA strand, and prevent loading of the passenger strand, as annotated in miRBase (www.mirbase.org). In some cases of miRNA deficiency, mimics can even be administered to restore miRNA expression level [286].
Notably, in major in vivo studies, transgenic knock-out (KO) or knock-in (KI) mice for a particular miRNA helped elucidate its function in organ specific manner for conditional transgene expression in both steady-state and pathologic-state [287,288]. However, beyond offering an understanding to miRNAs ’ functionality, synthetic miRNAs can offer novel potential therapeutic agents of great interest.
Delivery systems for miRNAs therapeutic purposes
The introduction of synthetic miRNAs into cells can be achieved using mainly lipid based transfection or electroporation parameters. For in vivo administration, this field remains challenging since miRNAs have to overcome many obstacles such as degradation by endogenous nucleases, renal clearance, ineffective uptake by target cells and activation of the immune defenses [289,290]. In addition, the strand selection and generation of mature strand from the same hairpin should be considered. Another major issue is the capacity of a same miRNA to affect multiple targets genes in different cell types and to regulate different mechanisms. So, the challenge is to deliver oligomiR in a tissue specific fashion to avoid unwanted side effects. Taking advantage of their tiny size, miRNAs can be encapsulated into nanoparticles to outflow the lipid barrier. Different delivery systems relying on liposomes delivery (neutral or cationic lipoplexes) or on polymers or on viral vectors exist with different pros and cons regarding toxicity, efficacy, stability, immunogenicity and safety [291-297]. Physical triggered delivery approaches may be also considered to deliver synthetic miRNAs [298]. They are interesting because they can confine the delivery of synthetic miRNAs in a specific area on demand. There is an excellent recent review that summarized the different strategies proposed so far for miRNA topical delivery of skin disorders [299]. Nonetheless, nude synthetic RNAs delivered without any vectors can be possible with limited cellular uptake and rapid degradation [300]. Nowadays, engineered miRNA therapeutics in preclinical stage has been developed (Table 2).
| Targeted miRNA | OligomiR | Application | Stage |
|---|---|---|---|
| miR-10b | AntagomiR | Glioblastoma | Preclinical |
| miR-221 | AntagomiR | Hepatocellular carcinoma | Preclinical |
| miR-21 | AntagomiR | Renal fibrosis | Preclinical |
| miR-33 | AntagomiR | Atherosclerosis | Completed preclinical |
| miR-122 | AntagomiR | Hepatitis C Virus infection | Phase IIa |
| miR-34 | Mimic | Primary liver cancer or solid cancers related to liver | Phase I |
| miR-155 | AntagomiR | Hematological malignancies | Completed preclinical |
| miR-92 | AntagomiR | Peripheral artery disease | Preclinical |
| miR-15 | AntagomiR | Myocardial infraction | Preclinical |
Table 2: miRNA therapeutics pre-clinical and clinical status adapted from [301].
Currently, two potential miRNAs based drugs are in phase I and phase II clinical trials. MRX34, a liposomal formulation of miR-34a mimic, is in phase I for patients with advanced solid tumors [301,302]. Miravirsen an miR-122 inhibitor is in phase II trial as a therapeutic agent for chronic Hepatitis C infection [303]. A Phase 1 clinical study was recently initiated for MRG-201, a miRNA mimic to microRNA-29b, as an antifibrotic agent for cutaneous and pulmonary fibrosis. These achievements for miRNA therapeutics give insight to their possible availability in the market in the upcoming years for the treatment of various diseases including chronic inflammatory diseases.
Nowadays, miRNAs are considered as key players in regulating molecular and cellular pathways of several physiological and pathological processes. They could be used either as diagnosis/ prognosis biomarkers or for therapeutic purposes. Indeed, they can be detected in small volume of blood or tissues samples. The miRNAs that are upregulated could be inhibited with miRNAs inhibitors whereas those that are downregulated can be supplemented with the delivery of miRNA mimics. As any nucleic acids, the delivery of those molecules is still challenging. Nevertheless, for skin applications the large variety of topical approaches made therapeutic miRNAs accessible for skin treatments. Since, one of miRNA features is its spatiotemporal expression; it is of importance to have a clear view on the kinetic of miRNA functionality to handle with precision their regulation. The challenge is to decipher the spatiotemporal miRNA deregulation. For skin diseases, mice models have contributed to important scientific outputs despite their limitations due to the anatomical differences between human and mice skin. To overcome such limitation, one could use human skin xenograft in nude mice. However, it is an expensive model and hard to handle. The best choice would be to build a human skin equivalent with a biomimetic organ on a chip device. In addition to be more close to human skin anatomy and microenvironement, such system could provide tools to follow up miRNAs expression as function of time and cell specific manner. Moreover, this device - through the high throughput and automation of culture - could offer the possibility to evaluate the synthetic miRNAs delivery and their functionality. Overall, the emerging miRNAs therapeutics is still under development and more studies must be done to validate their potentiality to supplement existing therapeutic approaches.
The authors declare no competing financial interest.
The work was supported by the grants from Région Centre, New Biomarkers-COSMETOSCIENCE ARD2020, Centre Val de loire, Cosmetic Valley and Centre Nationale de la recherche scientifique, Orléans, France.
Citation: Abdallah F, Pichon C (2019) MicroRNAs in skin biology: Biogenesis, Regulations and Functions in Homeostasis and Diseases. Immunome Res 15:169. doi: 10.35248/1745-7580.19.15.169
Received: 02-Jun-2019 Accepted: 25-Jul-2019 Published: 05-Aug-2019 , DOI: 10.35248/1745-7580.19.15.169
Copyright: © 2019 Abdallah F, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.