
Journal of Clinical Chemistry and Laboratory Medicine
Open Access
ISSN: 2736-6588

ISSN: 2736-6588
Research Article - (2024)Volume 7, Issue 1
Objective: Cardiac function stands as a robust and seemingly independent predictor of all-cause and cardiovascular mortality among individuals undergoing Hemodialysis (HD). The crucial need for efficient cardiac function assessment led us to explore the potential of using accessible blood sampling for evaluation. In this study, we cautiously harnessed cardiovascular proteomics in conjunction with Machine Learning (ML) techniques to explore the feasibility of predicting cardiac function in HD patients.
Methods: A cohort of 328 HD patients was gathered from two units located in Southern Taiwan. Utilizing proximity extension assays, a comprehensive measurement of 184 cardiovascular proteins was performed. Employing machine learning, we optimized a model for predicting cardiac dysfunction based on ejection fraction. Model performance was evaluated using the Area Under the Curve (AUC), while the Significance of Hierarchical Averaging of Shapley Values (SHAP) method was employed to identify crucial variables for prediction.
Results: Employing a dataset encompassing 184 proteomic biomarkers and 34 standard clinical variables within our analytical framework, it was discerned that the predictive efficacy of the "proteomic biomarkers" surpassed that of the "routine clinical and laboratory variables" using various machine learning algorithms, including Classification And Regression Tree (CART), Least Absolute Shrinkage And Selection Operator (LASSO), random forest, ranger and extreme gradient boosting (XgBoost) models. Through the application of XgBoost for feature selection, the significance of N-terminal pro-B type Natriuretic Peptide (NT-proBNP) emerged as the foremost contributor, supplemented by the predictive roles of Angiotensin Converting Enzyme 2 (ACE-2) and Chitotriosidase-1 (CHIT- 1) in determining cardiac dysfunction. This alignment was reaffirmed by SHAP-based elucidation of the XgBoost model.
Conclusion: Proteomic features outperformed clinical variables in predicting cardiac dysfunction using machine learning. Further analysis with XgBoost and SHAP highlighted NT-proBNP and CHIT-1 as crucial biomarkers, shedding light on cardiac dysfunction assessment in HD patients through blood biomarkers.
Cardiac function; Hemodialysis; Cardiovascular proteomic; Machine learning
Cardiovascular disease holds its status as a leading cause of mortality among patients with End-Stage Renal Disease (ESRD), with Congestive Heart Failure (CHF) prevailing at an approximate rate of 36-44% [1-3]. Post hemodialysis, there exists a yearly incidence of newly diagnosed CHF at 7%, which is accompanied by a relatively poorer survival rate [4,5]. Furthermore, cardiac function maintains its position as a robust predictor of all-cause and cardiovascular death in patients receiving Hemodialysis (HD) [6,7]. Besies, Left Ventricular (LV) dysfunction also accounts for a portion of Sudden Cardiac Death (SCD) in HD patients [8-10]. Given this context, the imperative to discover a more accessible method for the feasible evaluation of cardiac function in HD-dependent patients is paramount.
It is promising to evaluate cardiac function through circulating biomarkers because the blood sampling is less operator-independent and more cost-effective method in regular HD patients, compared to echocardiography. Many proteins, including plasma natriuretic peptides, high-sensitivity troponins and soluble suppression of tumorigenesis-2, have been introduced to in daily clinical practice as a diagnostic tool for CHF [11]. There are still lots of emerging biological markers including galectin-3, Human Epididymis protein 4 (HE4), Insulin-Like Growth Factor-Binding Protein 7 (IGFBP-7), Heart Fatty Acid-Binding Protein (H-FABP), Insulin-Like Growth Factor-Binding Protein 7 (IGFBP-7), Heart Fatty Acid-Binding Protein (H-FABP), soluble Cluster Of Differentiation 146 (sCD146), Interleukin-6 (IL-6), Growth Differentiation Factor 15 (GDF-15), Neutrophil Gelatinase-Associated Lipocalin (NGAL), Kidney Injury Molecule-1 (KIM-1) were mentioned to diagnose or stratify the prognosis [12-14]. However, prior investigations encountered constraints in measuring a restricted set of proteins and were not conducted within the HD population. The utilization of proximity extension assays, however, empowers the simultaneous measurement of an extensive spectrum of proteins, thus opening avenues for a plausible prediction model centered around markers obtained through blood sampling.
Assessing the performance of prediction models using various parameters necessitates the utilization of Machine Learning (ML) techniques to compare the prediction models established through the novel proteomic panel, electrocardiography parameters and clinical variables [15]. ML applications in CHF research have recently gained popularity and applied in various studies [16-18]. In this study, we aimed to predict cardiac dysfunction with machine learning strategies by using cardiovascular proteomics in HD patients. The outcomes of this research hold the potential to influence future clinical practices by providing a non-invasive and comprehensive method for evaluating cardiac function in HD patients, thus enabling timely interventions and improved patient outcomes.
Participants
From August 2016 through January 2017, we recruited 347 participants from two HD units (Kaohsiung Medical University Hospital and Kaohsiung Municipal Hsiao-Kang Hospital) in Southern Taiwan. Eligible participants were at least 30 years of age and receiving maintenance dialysis for at least 90 days. All patients received regular HD three times per week using automated volumetric machines. Each HD session lasted 3.5-4 hours and involved using high-flux dialyzers. The blood flow rate was controlled between 250 and 300 ml/min, the dialysate flow was maintained at 500 ml/min and the single pool Kt/V was more than 1.2 per week. The Institutional Review Board approved the study protocol of Kaohsiung Medical University (KMUHIRB-E(I)-20160095 and KMUHIRB-E(I)-20180139). Informed consent was obtained from all subjects.
Clinical parameters
The baseline characteristics of HD patients were recorded from electronic healthcare record systems, including age, sex, dialysis vintage, arteriovenous access type (fistula or graft), the primary cause of kidney failure (hypertension, diabetes, glomerulonephritis or others), comorbidities, medications and biochemical data. Hypertension was defined as blood pressure over 140/90 mmHg or taking blood pressure-lowering drugs. Diabetes mellitus was defined as Glycated hemoglobin (HbA1C) of over 6.5% or taking glucose-lowering drugs. The biochemical data were obtained from routine blood samples within 30 days before Cardiovascular (CV) proteomics measurement. We collected blood samples at the beginning of the week after overnight fasting from patients through the arteriovenous access immediately before the scheduled HD session and stored them at -80°C.
Echocardiography
The echocardiographic examination was performed using VIVID 7 by experienced cardiologists to assess cardiac structure and function. According to the American Society of Echocardiography recommendations, two-dimensional and two-dimensional guided M-mode images were recorded from standardized views. The Doppler sample volume was placed at the tips of the mitral leaflets to obtain the LV inflow waveforms from the apical four-chamber view. Pulsed Doppler tissue imaging was performed with the sample volume placed at the lateral and septal corners of the mitral annulus to obtain waveforms from the apical four-chamber view. Interventricular Septal Wall Thickness in diastole (IVSTd), Left Ventricular Internal Diameter in diastole (LVIDd) and Left Ventricular Posterior Wall Thickness (LVPWT) were measured in the left parasternal long-axis view. The Left Ventricular Ejection Fraction (LVEF) was measured using Simpson’s modified method.
Cardiovascular proteomics
The Proseek Multiplex 96 × 96 proximity extension assay simultaneously measured 184 proteins. In brief, two protein-specific antibodies attached to oligonucleotide strands were used. Each sample contained two incubations, one extension and one detection control to determine the lower detection limit and normalize the measurements. When both antibodies are bound to the target protein, the oligonucleotides are brought together and amplified in a quantitative Polymerase Chain Reaction (qPCR). The relative concentration of the target protein was correlated to the qPCR values. Normalized Protein Expression (NPX) values were generated from qPCR quantification cycle values by log2-transformation. The NPX values were corrected for technical variation by an interpolate control and determined limited detection via a negative control. Mean intra-assay and inter-assay coefficients of variation were 4% and 10%, respectively. Quality control was performed to remove proteins with >15% samples below the detection limit and subjects with a high proportion of missing protein values.
Study design and statistical analysis
The general workflow was summarized in Figure 1. In this study, we recruited 347 HD patients, incorporating measurements of 184 blood cardiovascular protein biomarkers and 34 clinical variables as features. Later, 19 participants were excluded due to missing Left Ventricular Ejection Fraction (LVEF) data, leaving 328 participants for the subsequent analysis. The objective was to predict a binary cardiac function status, categorized as 0 and 1 (preserved: 0, reduced: 1) based on different LVEF cutoff points. The dataset was divided into training and validation sets with an 8:2 ratios. Model parameter optimization was performed through 50-fold cross-validation on the training set. Following the 2023 Consensus of Taiwan Society of Cardiology, a left ventricular ejection fraction below 50% was considered indicative of reduced cardiac function, thus a threshold of 50% was used [19]. To validate the robustness of results, a sensitivity analysis was conducted with a 45% cutoff point. For model training, five machine learning classification algorithms were employed, including Classification and Regression Tree (CART), Least Absolute Shrinkage and Selection Operator (LASSO), random forest, Ranger optimizer and extreme gradient boosting (XgBoost). Evaluations were conducted using the Area Under the Curve (AUC) of the receiver operating characteristic, employing 5-fold cross-validation. The algorithm demonstrating the highest mean AUC across 50 iterations was identified as the optimal performer. Furthermore, Shapley Additive exPlanations were used to interpret feature influences on predictions within the model. The analytical procedures were carried out using R (version 4.1.1) and involved packages including rpart, randomForest, ranger, xgboost, glmnet and ggplot-2 for both data analysis and visualization.
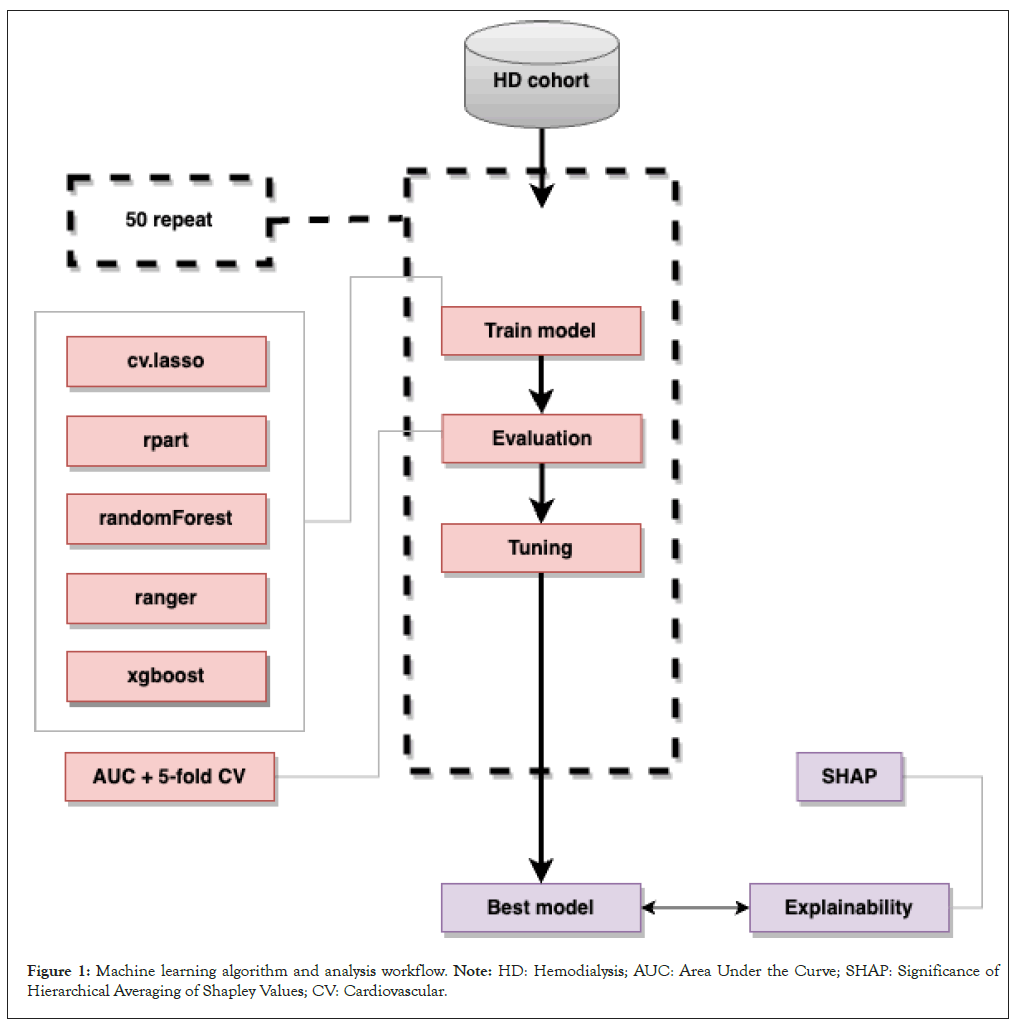
Figure 1: Machine learning algorithm and analysis workflow. Note: HD: Hemodialysis; AUC: Area Under the Curve; SHAP: Significance of Hierarchical Averaging of Shapley Values; CV: Cardiovascular.
Baseline characteristics
Within the cohort of 328 patients undergoing hemodialysis, the mean age was 59.16, accompanied by an average hemodialysis vintage of 7.34 years. Among these patients, 307 (93.6%) were classified as "preserved," while 21 (6.4%) were categorized as "reduced" using a LVEF cutoff of 50%. Similarly, when the LVEF cutoff was established at 45%, 314 (95.7%) were classified as "preserved" and 14 (4.3%) were categorized as "reduced." Between the reduced and preserved groups using a LVEF cutoff of 50%, most clinical and biochemical profiles were not differences except physical activity, hematocrit, Urea Removal Rate (URR) and Kt/V (Table 1). In similar, no differences in baseline characteristics except uric acid, parathyroid hormone and Kt/V between the reduced and preserved groups using a LVEF cutoff of 45%.
| Characteristics | LVEF ≥ 50% (n = 307) | LVEF <50% (n = 21) | P value |
|---|---|---|---|
| Age (year) | 58.98 ± 11.4 | 61.8 ± 11.7 | 0.274 |
| Male gender (%) | 163 (53%) | 15 (71%) | 0.103 |
| Smoking | 34 (11%) | 1 (5%) | 0.365 |
| Hypertension (%) | 238 (78%) | 17 (81%) | 0.715 |
| DM (%) | 129 (42%) | 11 (52%) | 0.353 |
| Coronary artery disease (%) | 53 (17%) | 7 (33%) | 0.065 |
| Cardiovascular disease (%) | 29 (9%) | 1 (5%) | 0.471 |
| Atrial fibrillation (%) | 14 (5%) | 2 (10%) | 0.307 |
| Malignancy (%) | 38 (12%) | 1 (5%) | 0.297 |
| Hyperlipidemia (%) | 118 (38%) | 9 (43%) | 0.687 |
| Liver cirrhosis (%) | 9 (3%) | 1 (5%) | 0.637 |
| Hepatitis B (%) | 39 (13%) | 3 (14%) | 0.834 |
| Hepatitis C (%) | 31 (10%) | 4 (19%) | 0.199 |
| Gout (%) | 38 (13%) | 4 (19%) | 0.376 |
| Parathyroidectomy (%) | 65 (21%) | 3 (14%) | 0.451 |
| Cause of ESRD | - | - | 0.387 |
| Hypertension | 34 (11.1%) | 1 (4.8%) | - |
| Diabetes mellitus | 106 (34.5%) | 10 (47.6%) | - |
| Glomerulonephritis | 113 (36.8%) | 5 (23.8%) | - |
| Others | 54 (17.6%) | 5 (23.8%) | - |
| Arteriovenous shunt type | - | - | 0.726 |
| Fistula (%) | 271 (88%) | 36 (86%) | - |
| Graft (%) | 36 (12%) | 3 (14%) | - |
| Physical activity | 82.57 ± 12.26 | 74.47 ± 12.97 | 0.004 |
| HD vintage (month) | 89.04 ± 69.17 | 74.76 ± 65.4 | 0.359 |
| Body weight before HD (Kg) | 62.94 ± 12.00 | 64.05 ± 12.09 | 0.682 |
| Body weight after HD (Kg) | 60.52 ± 11.61 | 61.65 ± 11.95 | 0.666 |
| Ultrafiltration (Kg) | 2.42 ± 0.96 | 2.40 ± 0.93 | 0.896 |
| SBP (mmHg) | 146.12 ± 25.17 | 143.20 ± 22.80 | 0.605 |
| DBP (mmHg) | 79.67 ± 13.62 | 76.20 ± 16.17 | 0.265 |
| Body mass index (kg/m²) | 23.65 ± 3.74 | 24.10 ± 3.83 | 0.591 |
| Height (cm) | 162.27 ± 7.75 | 162.96 ± 6.04 | 0.689 |
| Body weight (kg) | 62.45 ± 11.69 | 64.11 ± 11.06 | 0.528 |
| Laboratory parameters | - | - | - |
| WBC (10³/μL) | 6.21 ± 1.90 | 5.91 ± 1.35 | 0.479 |
| RBC (106/μL) | 3.65 ± 0.59 | 3.52 ± 0.72 | 0.352 |
| Hemoglobin (g/dL) | 10.68 ± 1.17 | 10.45 ± 1.64 | 0.531 |
| Hematocrit (%) | 33.2 ± 3.84 | 31.41 ± 4.85 | 0.042 |
| MCV (fL) | 91.32 ± 7.54 | 92.28 ± 7.26 | 0.057 |
| Platelet (10³/μL) | 189.48 ± 60.0 | 183.76 ± 54.92 | 0.672 |
| Total protein (g/dL) | 6.88 ± 0.54 | 6.91 ± 0.42 | 0.795 |
| Albumin (g/dL) | 3.86 ± 0.28 | 3.85 ± 0.34 | 0.86 |
| AST (U/L) | 17.35 ± 10.00 | 16.78 ± 8.34 | 0.799 |
| ALT (U/L) | 14.41 ± 9.41 | 12.78 ± 6.85 | 0.435 |
| ALP (U/L) | 237.04 ± 205.13 | 295.53 ± 505.59 | 0.27 |
| Total cholesterol (mg/dL) | 168.98 ± 40.24 | 154.66 ± 36.03 | 0.113 |
| Triglyceride (mg/dL) | 150.13 ± 96.00 | 138.77 ± 71.53 | 0.595 |
| LDL-cholesterol (mg/dL) | 95.46 ± 32.99 | 83.53 ± 24.12 | 0.104 |
| HDL-cholesterol (mg/dL) | 41.39 ± 12.17 | 40.96 ± 10.67 | 0.874 |
| Fasting glucose (mg/dL) | 112.23 ± 49.38 | 109.94 ± 29.75 | 0.834 |
| BUN (mg/dL) | 66.08 ± 14.20 | 64.22 ± 14.71 | 0.562 |
| Creatinine (mg/dL) | 10.2 ± 2.08 | 9.52 ± 2.18 | 0.144 |
| Uric acid (mg/dL) | 7.56 ± 1.47 | 6.95 ± 1.58 | 0.069 |
| Sodium (mmol/L) | 138.43 ± 3.25 | 137.57 ± 2.43 | 0.234 |
| Potassium (mmol/L) | 4.58 ± 0.63 | 4.50 ± 0.61 | 0.603 |
| Total calcium (mg/dL) | 9.28 ± 0.92 | 9.26 ± 0.71 | 0.906 |
| Phosphate (mg/dL) | 4.72 ± 1.11 | 4.94 ± 1.12 | 0.383 |
| URR | 0.72 ± 0.05 | 0.69 ± 0.04 | 0.014 |
| Kt/V | 1.57 ± 0.24 | 1.43 ± 0.17 | 0.013 |
| nPCR | 1.08 ± 0.20 | 1.08 ± 0.21 | 0.903 |
| Fe (μg/dL) | 62.83 ± 22.68 | 63.25 ± 19.8 | 0.933 |
| UIBC (μg/dL) | 143.02 ± 44.96 | 141.70 ±42.62 | 0.896 |
| Ferritin (ng/mL) | 464.44 ± 339.35 | 569.33 ± 243.54 | 0.165 |
| Transferrin (mg/dL) | 31.36 ± 12.21 | 31.42 ± 9.54 | 0.981 |
| Aluminum (μg/L) | 20.86 ± 14.84 | 18.79 ± 13.6 | 0.535 |
| Magnesium (mg/dL) | 2.63 ± 0.42 | 2.56 ± 0.34 | 0.471 |
| Zinc (μg/dL) | 96.90 ± 17.52 | 93.93 ± 13.00 | 0.446 |
| PTH (pg/mL) | 403.67 ± 322.71 | 594.07 ± 491.45 | 0.095 |
| CRP (mg/L) | 2.17 ± 3.66 | 3.28 ± 5.64 | 0.199 |
Note: DM: Diabetes Mellitus; ESRD: End-Stage Renal Disease; HD: Hemodialysis, SBP: Systolic Blood Pressure; DBP: Diastolic Blood Pressure; WBC: White Blood Cell; RBC: Red Blood Cell; MCV: Mean Corpuscular Volume; AST: Aspartate Aminotransferase; ALT: Alanine Aminotransferase; ALP: Alkaline Phosphatase; BUN: Blood Urea Nitrogen; HDL: High-Density Lipoprotein; LDL: Low-Density Lipoprotein; URR: Urea Removal Rate; nPCR: Normalized Protein Catabolic Rate; UIBC: Unsaturated Iron Binding Capacity; PTH: Parathyroid Hormone; CRP: C-Reactive Protein.
Table 1: Baseline characteristics of routine clinical and laboratory variables in hemodialysis patients according to the Left Ventricular Ejection Fraction (LVEF) group (LVEF ≥ 50% and LVEF <50%).
Comparison of the efficacy of prediction with different features
In the pursuit of identifying the most optimal prediction model, we conducted a comparative analysis between the predictive capacities of a set of clinical and laboratory variables against 184 proteins, utilizing machine learning algorithms to determine the binomial outcome of reduced or preserved heart function. When using a LVEF cutoff of 50%, the weighted model demonstrated that proteomic biomarkers outperformed routine clinical variables in terms of prediction efficacy, as depicted in Figure 2A. This trend persisted when the LVEF cutoff was set at 45%, as seen in Figure 2B. Similar results were presented in the unweighted model.
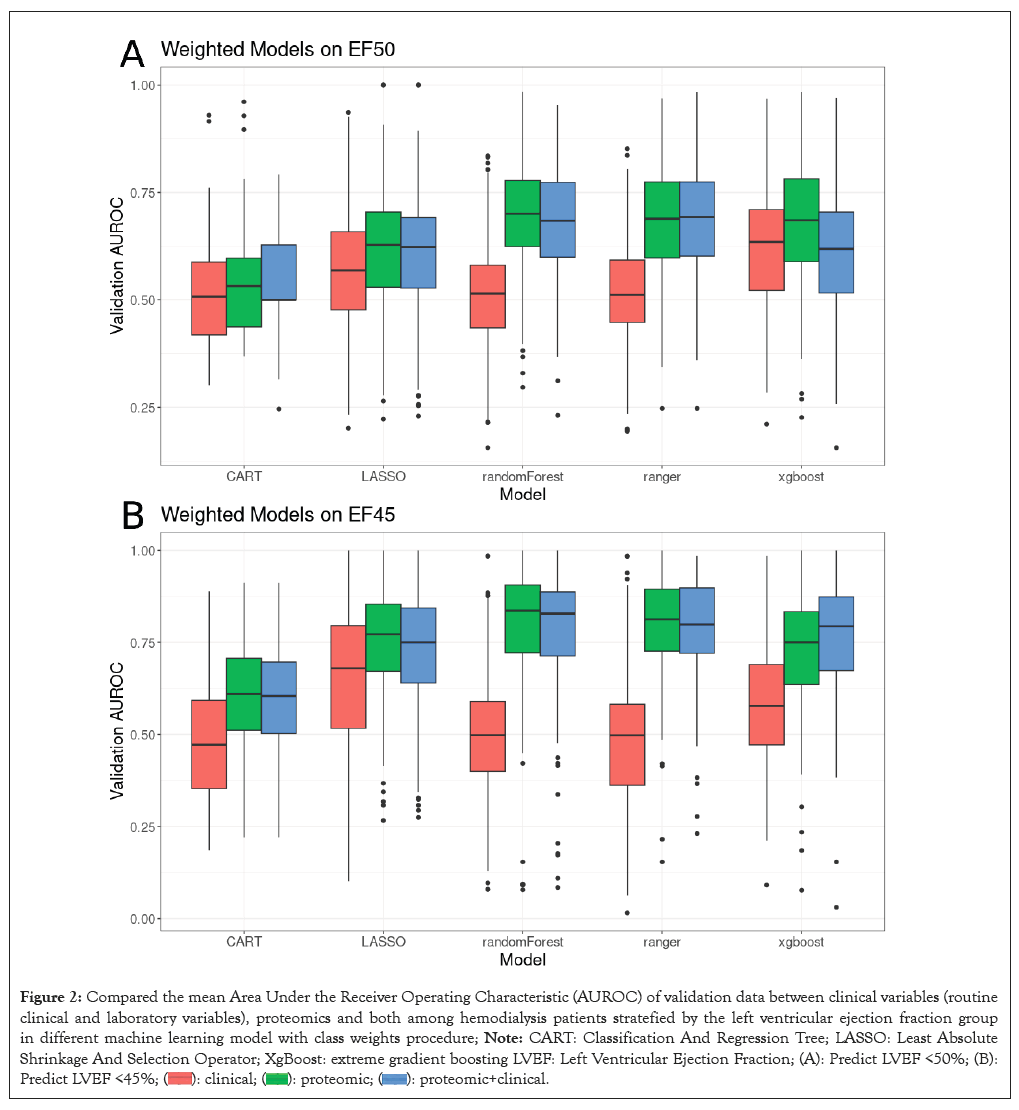
Figure 2: Compared the mean Area Under the Receiver Operating Characteristic (AUROC) of validation data between clinical variables (routine
clinical and laboratory variables), proteomics and both among hemodialysis patients stratefied by the left ventricular ejection fraction group
in different machine learning model with class weights procedure; Note: CART: Classification And Regression Tree; LASSO: Least Absolute
Shrinkage And Selection Operator; XgBoost: extreme gradient boosting LVEF: Left Ventricular Ejection Fraction; (A): Predict LVEF <50%; (B):
Predict LVEF <45%; 
Interpretation of XGBoost model feature importance using significance of hierarchical averaging of shapley values
Since cardiovascular proteomics provide a better prediction for cardiac dysfunction than clinical and laboratory variables, we performed the model explanation by SHAP to explore the import protein biomarkers. The SHAP value analysis provided insights into the feature importance as determined by the predictive model. Two separate plots illustrate the distribution of SHAP values for cardiac dysfunction prediction by LVEF less than 50% (Figure 3A) or less than 45% (Figure 3B).
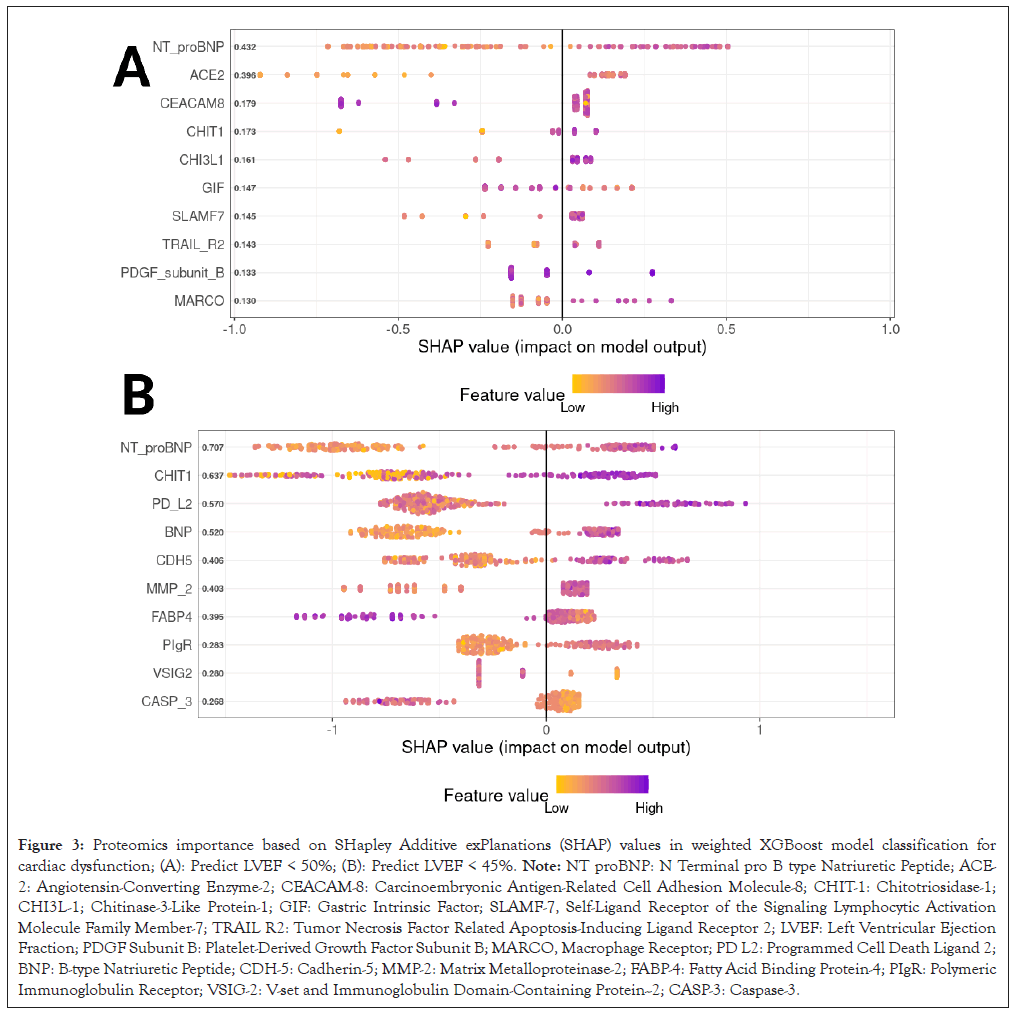
Figure 3: Proteomics importance based on SHapley Additive exPlanations (SHAP) values in weighted XGBoost model classification for cardiac dysfunction; (A): Predict LVEF < 50%; (B): Predict LVEF < 45%. Note: NT proBNP: N Terminal pro B type Natriuretic Peptide; ACE- 2: Angiotensin-Converting Enzyme-2; CEACAM-8: Carcinoembryonic Antigen-Related Cell Adhesion Molecule-8; CHIT-1: Chitotriosidase-1; CHI3L-1; Chitinase-3-Like Protein-1; GIF: Gastric Intrinsic Factor; SLAMF-7, Self-Ligand Receptor of the Signaling Lymphocytic Activation Molecule Family Member-7; TRAIL R2: Tumor Necrosis Factor Related Apoptosis-Inducing Ligand Receptor 2; LVEF: Left Ventricular Ejection Fraction; PDGF Subunit B: Platelet-Derived Growth Factor Subunit B; MARCO, Macrophage Receptor; PD L2: Programmed Cell Death Ligand 2; BNP: B-type Natriuretic Peptide; CDH-5: Cadherin-5; MMP-2: Matrix Metalloproteinase-2; FABP-4: Fatty Acid Binding Protein-4; PIgR: Polymeric Immunoglobulin Receptor; VSIG-2: V-set and Immunoglobulin Domain-Containing Protein--2; CASP-3: Caspase-3.
In Figure 3A, NT proBNP emerged as the feature with the highest median SHAP value, indicating its strong positive impact on the model's output. This was followed by features such as Angiotensin-Converting Enzyme-2 (ACE-2) and Carcinoembryonic Antigen-Related Cell Adhesion Molecule 8 (CEACAM8). The color intensity on the plot reflects the magnitude of the feature values, with the distribution of SHAP values for ACE-2 and CEACAM8 revealing a mix of low and high feature values across the dataset. Figure 3B also highlighted NT proBNP as a significant feature, following by Chitotriosidase-1 (CHIT-1) and Programmed Cell Death 1 ligand 2 (PD L2), showing substantial variability in their SHAP values and thereby suggesting differential impacts on the model prediction across the dataset instances. The consistency of NT proBNP as a top contributing feature in both cardiac dysfunction definition underscores its predictive importance.
Feature importance in predicting reduced left ventricular ejection fraction in XGBoost model
The feature importance for predicting LVEF below specific thresholds was quantitatively assessed, as depicted in Figure 4A and 4B. The analysis was bifurcated into two distinct predictive scenarios: LVEF less than 50% (Figure 4A) and LVEF less than 45% (Figure 4B).
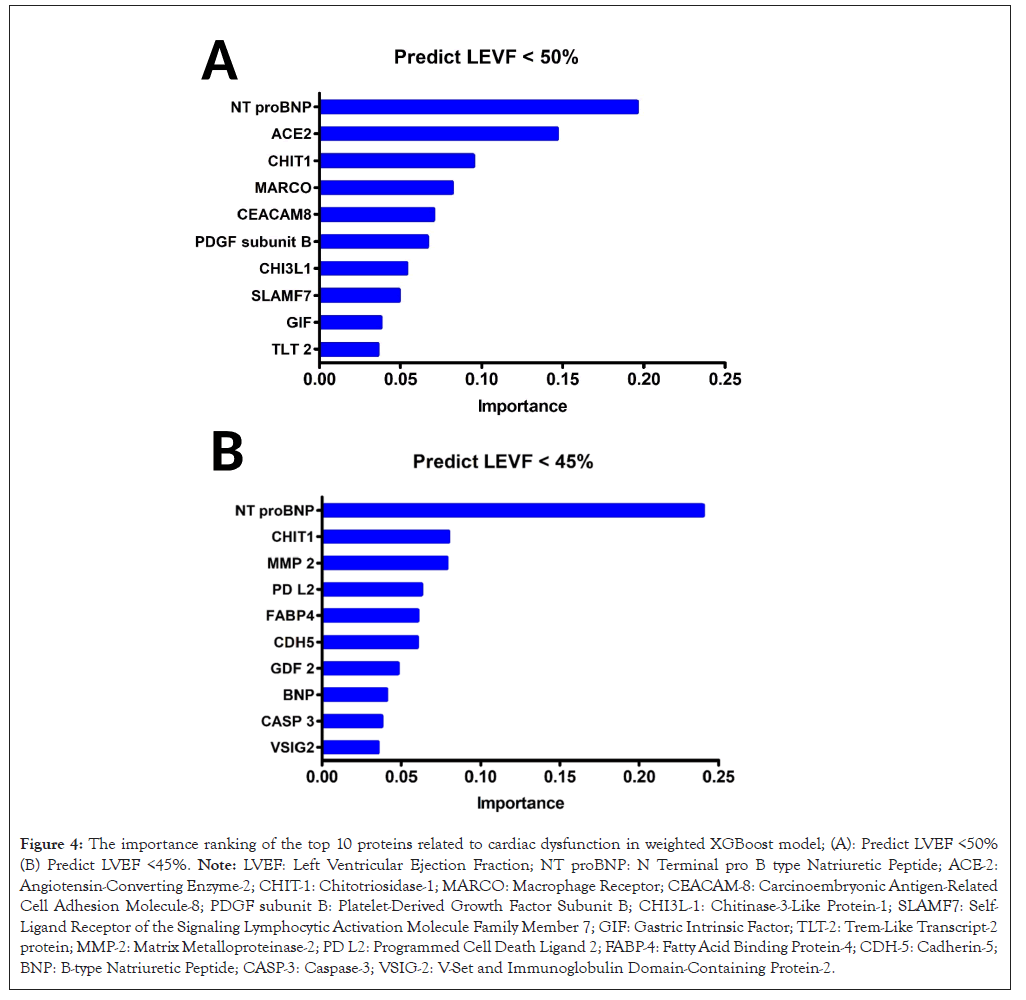
Figure 4: The importance ranking of the top 10 proteins related to cardiac dysfunction in weighted XGBoost model; (A): Predict LVEF <50% (B) Predict LVEF <45%. Note: LVEF: Left Ventricular Ejection Fraction; NT proBNP: N Terminal pro B type Natriuretic Peptide; ACE-2: Angiotensin-Converting Enzyme-2; CHIT-1: Chitotriosidase-1; MARCO: Macrophage Receptor; CEACAM-8: Carcinoembryonic Antigen-Related Cell Adhesion Molecule-8; PDGF subunit B: Platelet-Derived Growth Factor Subunit B; CHI3L-1: Chitinase-3-Like Protein-1; SLAMF7: Self- Ligand Receptor of the Signaling Lymphocytic Activation Molecule Family Member 7; GIF: Gastric Intrinsic Factor; TLT-2: Trem-Like Transcript-2 protein; MMP-2: Matrix Metalloproteinase-2; PD L2: Programmed Cell Death Ligand 2; FABP-4: Fatty Acid Binding Protein-4; CDH-5: Cadherin-5; BNP: B-type Natriuretic Peptide; CASP-3: Caspase-3; VSIG-2: V-Set and Immunoglobulin Domain-Containing Protein-2.
For the prediction of LVEF below the 50% threshold, the feature importance profile was led by NT-proBNP and. subsequent features including ACE-2 and CHIT-1 (Figure 4A). Additional features such as Macrophage Receptor with Collagenous Structure (MARCO) and CEACAM8 also contributed to the model, albeit with diminishing influence. When the threshold for LVEF was adjusted to below 45%, NT-proBNP consistently exhibited the highest feature importance, indicative of its robust association with cardiac functional impairment. Interestingly, the descending order of feature importance showed variations, with CHIT-1 and Matrix Metalloproteinase-2 (MMP-2) ascending in their respective ranks compared to the previous threshold model (Figure 4B). This shift suggests a dynamic interplay between the features and the degree of LVEF reduction, potentially reflecting the complex biological processes underlying severe cardiac dysfunction.
Across both thresholds, the unchanging prominence of NT-proBNP underscores its pivotal role as a biomarker in cardiac health assessment. The alteration in the rank and importance of other features between the two models potentially reveals the multifactorial nature of cardiac impairment, as well as the sensitivity of the predictive model to the severity of LVEF reduction. These findings illuminate the intricacies of feature interactions within the pathological spectrum of LVEF compromise and underscore the utility of machine learning approaches in elucidating the prognostic landscape of cardiac dysfunction in HD patients.
Utilizing machine learning classification algorithms, our inquiry substantiated that the ensemble of 184 proteins displayed augmented predictive efficacy pertaining to cardiac function, as compared to clinical variables, within the cohort of Hemodialysis (HD) patients. Furthermore, our analytical identified pivotal proteins-NT-proBNP, CHIT-1, ACE-2 and MMP-2 in the prediction of cardiac dysfunction. We use SHAP value, which explains the determinants behind machine learning model outputs by game theoretic approach, to depicts the feature importance and relationship between prediction results. In view of feature rank, NT-proBNP and CHIT-1 are highly correlated with reduced cardiac dysfunction in whether cut-off point of LVEF. However, ACE-2 predict cardiac dysfunction well in patients with LVEF less than 50%, but the importance is less in patients with LVEF LVEF less than 45%. These outcomes collectively underscore the promise intrinsic to the comprehensive application of proteomic methodologies in advancing cardiac dysfunction prediction, with potential implications for refining risk assessment and engendering targeted interventions, thus ameliorating the quality of patient care.
The data from the cardiovascular proteomic panel contains numerous results, so they are difficult to analysis through traditional statistic method. The ML technique provides a new approach to solve these complex data [20]. ML aims to increase prediction accuracy for certain task, while the traditional statistic method emphasizes the inference of relationship between variables and has been introduced into the field to diagnose congestive heart failure [21]. In previous studies, they use characterized unsupervised learning methods for subgroup identification and pathway analysis on pathophysiology [22-24]. In the present study, supervised learning strategy was used to interpret the collection data and hope to improve both quality and efficiency in real world clinical practice of HD healthcare.
Considering the hierarchy of feature ranking, NT-proBNP emerged as the primary feature within both the LASSO and XgBoost models. This outcome aligns consistently with prior research wherein NT-proBNP holds recommendations for the evaluation of cardiac function and diagnosis of congestive heart failure in international guidelines [25,26]. Its synthesis by ventricle myocytes in response to stretching stimuli and its role as a compensatory mechanism against volume overload by counteracting the kidney's renin-angiotensin-aldosterone system underscore its physiological importance [27-29]. Moreover, the correlation of NT-proBNP with left ventricle hypertrophy and systolic dysfunction enhances its multifaceted utility [30]. The CHIT-1 exhibited notable significance, particularly within the XgBoost model, across various heart function cut-points. CHIT-1 is predominantly secreted by human macrophages and plays a role in the immune processes contributing to atherosclerotic plaque formation in HD patients [31]. Additionally, its connection to the onset of heart failure raises the prospect of its potential as an important cardiac function biomarker, pending further clinical evidence [32]. The ACE-2 and MMP-2 surfaced as features of variable importance across diverse cardiac function cut-points within our feature selection analysis. ACE-2's vasodilatory and anti-proliferative actions through angiotensin II degradation, which counteracts the renin-angiotensin system, coincide with its augmented expression in heart failure contexts [33,34]. Elevated ACE-2 levels have demonstrated associations with incident heart failure and cardiovascular mortality, reinforcing its prognostic significance. Similarly, MMP-2's connections to heart failure and left ventricular remodelling are documented, accompanied by its implication in adverse cardiovascular events, further establishing its role in cardiac function prediction [35-37].
The study's strengths lie in its innovative approach, integrating a large, well-characterized proteomic panel covering 184 cardiovascular protein biomarkers with machine learning to predict cardiac dysfunction in HD patients. Application of multiple sophisticated machine learning algorithms for classification and comparison of predictive capacity. This rigorous analytical approach enhanced validity and generalizability of the findings. Use of model interpretation with SHAP values to systematically determine feature importance ranks and illustrate directionality of effects on predicting ejection fraction categories. This enhanced interpretability of the complex machine learning outputs. Besides, stratification of ejection fraction thresholds allowed assessment of consistency and variability across differing severities of cardiac functional impairment. This improved biological and clinical insights into the cardiac dysfunction spectrum. However, this study has several limitations that should be acknowledged. First, the sample size was relatively modest at 328 hemodialysis patients from two centres in Southern Taiwan. Our findings will require external validation in larger, more diverse patient cohorts to ensure generalizability. Second, this was a cross-sectional analysis, therefore we cannot confirm causal relationships or make inferences about long-term prognostic utility of the proteomic biomarkers. Longitudinal studies are warranted to determine how well the identified proteins predict future cardiac events and mortality. Third, our echocardiographic measurements were based on one-time assessments of cardiac structure and function. Serial imaging data could not be incorporated to account for variability over time. Additionally, we utilized left ventricular ejection fraction cut-offs to categorize preserved versus reduced cardiac function. Other echocardiographic parameters beyond ejection fraction may provide complementary information. Finally, the pathophysiologic mechanisms underlying the predictive proteins we identified have not been definitively characterized. Further experimental research is needed to elucidate the functional roles of CHIT-1 in the setting of cardiac injury and remodeling related to hemodialysis. Despite these limitations, our study takes an important step toward risk stratification and earlier identification of cardiac dysfunction in a high-risk population using an emerging proteomic approach.
Cardiovascular proteomic features demonstrate superior prediction for cardiac function status compared with clinical variables based on machine learning classification algorithms. The outcomes underscore the significance of both NT-proBNP and CHIT-1 as substantial proteomic biomarkers in the prediction of heart function, irrespective of the chosen LVEF cut-point. The study underscores the potential of utilizing a comprehensive proteomic approach to enhance the prediction of cardiac function, shedding light on the vital role of specific proteins in this context. These findings hold promise for refining risk assessment and potentially facilitating targeted interventions for improved patient care.
The funding sources did not play any role in the design or conduct of the study, collection, management, analysis, interpretation of the data or preparation, review or approval of the manuscript. The study was funded by grants from the Ministry of Science and Technology, Taiwan (MOST 107-2314-B-037-021-MY2, MOST 109-2314-B-037-102-MY2 and MOST 111-2314-B-037 -083 -MY3), Kaohsiung Medical University Hospital, Taiwan (KMUH109-9R17, KMUH110-0R19, KMUH111-1R14, KMUH110-0M73, KMUH111-1R73, KMUH111-1M60 and KMUH-DK(B)110003-4), Kaohsiung Medical University, Taiwan (NHRIKMU-111-I003-1 and NHRIKMU-111-I003-2) and NSYSU-KMU JOINT RESEARCH PROJECT (NSYSUKMU 111-P21).
Conceptualization, Jen-Ping Lee, Yu-Lin Chao, Ping-Hsun Wu, Yi-Ting Lin, Yi-Wen Chiu and Mei-Chuan Kuo; Data curation, Jen-Ping Lee, Yu-Lin Chao, Chan Hsu, Ping-Hsun Wu and Yi-Ting Lin; Study design and analysis plan, Jen-Ping Lee, Yu-Lin Chao, Ping-Hsun Wu, Chan Hsu and Yi-Ting Lin; Statistical analysis, Jen-Ping Lee, Yu-Lin Chao and Chan Hsu; Funding acquisition, Ping-Hsun Wu, Yi-Ting Lin and Mei-Chuan Kuo; Investigation, Ping-Hsun Wu, Pei-Yu Wu, Szu-Chia Chen, Wei-Chung Tsai, Yi-Wen Chiu, Shang-Jyh Hwang, Yi-Ting Lin and Mei-Chuan Kuo; Writing-first draft, Jen-Ping Lee, , Yu-Lin Chao, Ping-Hsun Wu, Yi-Ting Lin; Writing-review & editing, Jen-Ping Lee, Yu-Lin Chao, Ping-Hsun Wu, Yun-Shiuan Chuang, Pei-Yu Wu, Szu-Chia Chen, Wei-Chung Tsai, Yi-Wen Chiu, Shang-Jyh Hwang, Yi-Ting Lin and Mei-Chuan Kuo.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Lee JP, Chao YL, Wu PH, Chuang YS, Hsu C, Wu PY, et al. (2024) Machine Learning-Based Prediction of Cardiac Dysfunction in Hemodialysis Patients through Blood Cardiovascular Proteomics. J Clin Chem Lab Med. 7:283.
Received: 23-Jan-2024, Manuscript No. JCCLM-24-29303; Editor assigned: 25-Jan-2024, Pre QC No. JCCLM-24-29303 (PQ); Reviewed: 08-Feb-2024, QC No. JCCLM-24-29303; Revised: 15-Feb-2024, Manuscript No. JCCLM-24-29303 (R); Published: 22-Feb-2024 , DOI: 10.35248/2736-6588.24.7.283
Copyright: © 2024 Lee JP, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.