Journal of Thermodynamics & Catalysis
Open Access
ISSN: 2157-7544
ISSN: 2157-7544
Research Article - (2015) Volume 6, Issue 1
When a physical system is complex it may possess a behaviour that displays some similarities with true living systems. If we consider, for instance, an enzyme reaction involving the random binding of two substrates A and B to the active site of the enzyme, the correct manner to study this reaction is to derive the rate equation as a combination of elements in interaction, namely the free enzyme E and the two enzyme-substrate complexes, EA and EB. In addition to these steady state concentrations there are local circuits of E, EA and EB. These circuits can be modified by the global system itself in such a way that, for pure physical reasons, the system can favour, for instance, a circuit running to E, to EA or to EB. These circuits exist and their functioning can be modulated by the system considered as a whole.
Keywords: Local reaction circuits; Multimolecular systems
Multimolecular systems can spontaneously change their topology according to certain physical laws. Such a situation could be considered a model for the self-organization of prebiotic systems that would not be “living” structures but could possibly possess some properties of living systems. An enzyme catalysed reaction can be considered as a network of enzyme states that mutually interact according to a certain topology, in such a way that the global system possesses some features of living systems. Such situations occur even with relatively simple enzyme systems in such a way that these systems seem to mimic primitive biological processes. As a matter of fact, it is quite possible that life has not appeared at once in multi molecular systems but has been preceded by a number of prebiotic systems that possessed some properties of true living systems.
The reasoning that has been followed in this Section can be extended to more complex and realistic situations involving the random binding of two, or several, ligands A and B to an enzyme [1]. As it is assumed that ligand binding takes place randomly, the overall process can be described as shown in Figure 1.
The mathematical treatment of the system upon pseudoequilibrium conditions leads to the classical equation
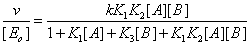 (1)
(1)
Where, as usual, v is the steady state reaction velocity of the process, [ Eo] the total enzyme concentration, [A] and [B] the substrate concentrations, K1 ,K2 and K3 the corresponding equilibrium binding constants, k the catalytic rate constant. Even though this equation fits the steady state rate data, it is not correct and should be considered as an approximation. As a matter of fact, such an enzyme system cannot be in equilibrium and at the same time catalyse a chemical process [2]. Hence this equation that usually fits the experimental data can only be considered as an approximation.
One should expect that the concentration of the various enzyme forms should be dependent upon the various local reaction flows that appear in the system during conversion of the substrate into the products. These reaction flows are basically dependent upon two types of events: substrate binding to and desorption from the enzyme on one hand, catalysis on the other hand [3]. Generation of the free enzyme E through substrate desorption is shown in Figure 2. The overall process involves substrate desorption from EA and EB states. The corresponding expression of the process leading to the free enzyme is then
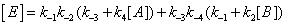 (2)
(2)
If we consider now the formation by desorption of state EA from the other states (Figure 3) the situation is now similar, although with some additional complexities, to the one previously described. For instance, the process described in states 3 and 4 (Figure 3) is similar, with some additional subtleties, to the one previously described. The processes in states 3 and 4 (Figure 3) can be written as
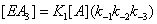 (3)
(3)
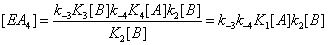 (4)
(4)
Hence the sum 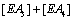 becomes
becomes
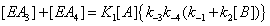 (4)
(4)
and the sum of the four terms leading to [EA] is now
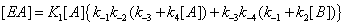 (5)
(5)
The same reasoning can be followed for the enzyme sates that bind substrate B instead of A. Thus the process in states 1 and 2 can be expressed as
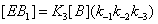 (6)
(6)
And
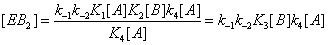 (7)
(7)
Hence the sum of 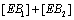 can be expressed as
can be expressed as
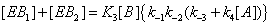 (8)
(8)
and the sum of the EB over the four states is now (Figure 4)
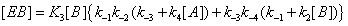 (9)
(9)
If now the enzyme has bound both A and B, the ternary EAB state can be generated through the process of Figure 3. Under these conditions one has
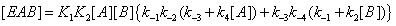 (10)
(10)
One can notice that, in this expression, the product K1 K2 equals that of K3 K4 . This is a consequence of the first principle of thermodynamics. It is important to stress that, in this reasoning, the concentrations of E,EA,EB and EAB are solely obtained through substrate desorption from the enzyme without taking account of the catalytic process (Figure 5).
The common dynamic process that we have found in the genesis of E, EA, EB, EAB and which is expressed by relation (17) and (18) can be described by the flows shown in Figures 6-8.
In the processes we have considered so far we have implicitly considered that E, EA and EB are generated through events that do not involve the catalytic constant k. There is no doubt; however, that catalytic constant k is, in fact, involved in the processes leading to the reappearance of E, EA and EB during the catalytic reaction. The steps leading to E and involving the catalytic constant k are shown in Figure 7. The corresponding sum of the flows involving rate constant k is
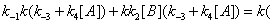
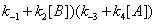 (11)
(11)
Similarly, the flows leading to EA and to EB are shown in Figures 7,8.
The mathematical expressions of the local flows uEA and uEBare
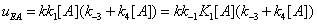 (12)
(12)
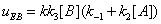
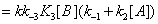
and they are described in Figures 7,8. Similarly the mathematical expression of the flow E u is (equation 11)
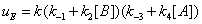 (13)
(13)
The concentrations of E, EA, EB and EAB can then be expressed as
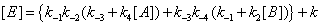
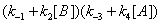
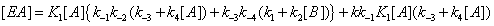 (14)
(14)
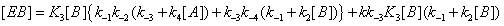
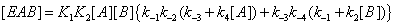
If we write these expressions relative to Enc , the free enzyme resulting from substrate desorption i.e.
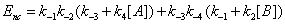 (15)
(15)
One has
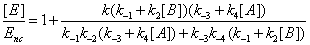
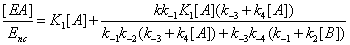 (16)
(16)
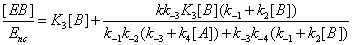
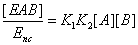
The u's that appear in these expressions are called the “relative local reactions flows”. They are defined as
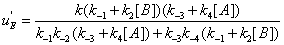
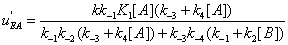 (17)
(17)
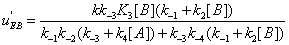
The u’s defined in expressions (23) represent the “relative reaction flows” involving the catalytic step and running to the free enzyme ) 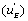 the EA complex
the EA complex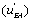 or the EB complex
or the EB complex 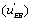 . The reaction flows are expressed relative to the substrate desorption leading to the free enzyme.
. The reaction flows are expressed relative to the substrate desorption leading to the free enzyme.
Let us consider now an enzyme reaction involving two substrates A and B, the corresponding steady sate rate equation assumes the form
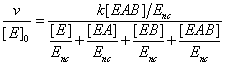 (18)
(18)
Moreover one has
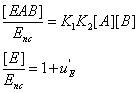
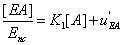
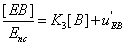 (19)
(19)
Hence equation (18) can be rewritten as
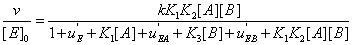 (20)
(20)
The same reasoning can be applied to probabilities and lead to similar results, for instance
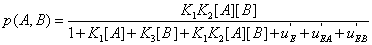 (21)
(21)
or p(A) , namely
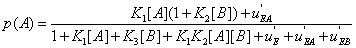 (22)
(22)
In rate equation (20), or in probabilities (21), (22) the u' appear in the denominator only, or both in the numerator and the denominator. Moreover in rate equation (20) it appears that, owing to the u' values, the catalytic constant k appears both in the numerator and the denominator of equations (22)
Let us consider, for instance, a protein that binds two ligands, A and B, randomly. In the absence of any reaction between A and B, or in the presence of a slow reaction between these ligands in such a way that the system is in equilibrium, or close to an equilibrium, the probability that the protein has bound a ligand [4], A for instance, is
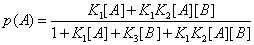 (22)
(22)
In this expression K1 ,K2 ,K3 are equilibrium binding constants of A and B to the protein. But one can define another probability, the probability, for instance, that the protein binds A after it has already bound B. One can define this conditional probability as
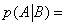
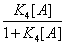 (24)
(24)
One can then define from these two probabilities, p(A) and 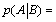 , an information, consumed or generated, i.e.
, an information, consumed or generated, i.e.
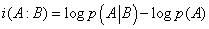 (25)
(25)
If i(A: B) > 0 , the interaction between A and B generates information, and conversely it consumes information if i(A: B) < 0 . These results are indeed valid for stable systems, i.e. for systems that do not evolve with time. But if the concentrations of A and B vary, expression (23) becomes
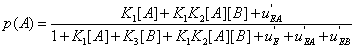 (26)
(26)
and the corresponding expression of 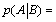 assumes the form
assumes the form
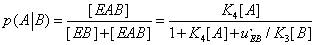 (27)
(27)
or
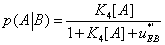 (28)
(28)
with 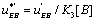 The expression of
The expression of 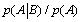 can be written as
can be written as
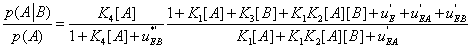 (29)
(29)
equivalent to
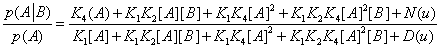 (30)
(30)
One can note that no emergence can appear in such a system if 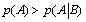 which means that
which means that 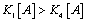 and
and 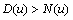 . The second condition is of particular interest for it shows whether or not the local reaction flows can generate information in the global system. One can write
. The second condition is of particular interest for it shows whether or not the local reaction flows can generate information in the global system. One can write
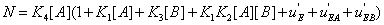
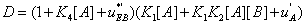 (31)
(31)
and the difference N-D assumes the form
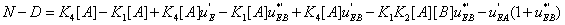 (32)
(32)
One can notice that the functions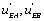 and
and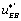 can be expressed as a function of
can be expressed as a function of 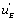 . One finds
. One finds
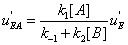
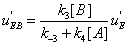 (33)
(33)
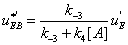
It follows that
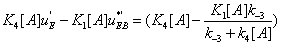 (34)
(34)
Similarly one has
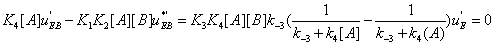 (35)
(35)
Hence the expression of N − Dis
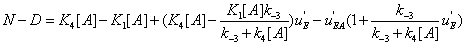 (36)
(36)
It follows from these expressions, which can be positive or negative, that the difference N − D can also be positive or negative. Hence one can conclude that the system can consume, or generate information.
In classical studies of enzyme kinetics it is implicitly, or explicitly, considered that the conversion of EAB into EB, EA and E are generated by simple desorption of the substrates A and B from the enzyme’s active site. What the present studies have shown is that the situation is far more complex. Regeneration of E, EA and EB is, to a certain extent, effected by simple desorption of the substrates A and B from the enzyme. But the present studies have also shown the existence of a far more complex system in which the regeneration of E, EA and EB is dependent upon circuits, or functions, we have called 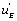 ,
, if a local circuit possesses the term
if a local circuit possesses the term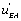 this means that this circuit is leading to the EA state. If, conversely, the local circuit contains the function
this means that this circuit is leading to the EA state. If, conversely, the local circuit contains the function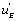 , this will favour the orientation of the flow toward the EB state. Last, but not least, if the local flow, or circuit, contains the
, this will favour the orientation of the flow toward the EB state. Last, but not least, if the local flow, or circuit, contains the or the
or the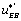 , term this will tend to orient the flow toward the E state.
, term this will tend to orient the flow toward the E state.
Another interesting result which is offered by the present mathematical analysis is that if 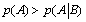 ) then
) then 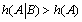 and the system generates some information. One can notice that depending on the sign of '
and the system generates some information. One can notice that depending on the sign of ' 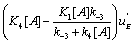 and K 4[A] − K1 [A] then states EB or EA predominate. Hence, in the first case, the “
and K 4[A] − K1 [A] then states EB or EA predominate. Hence, in the first case, the “ 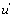 flow” is active and in the second case it is
flow” is active and in the second case it is 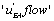 which is more active. All these results are consistent with the view that multimolecular networks are highly organized structures that can spontaneously generate this organization. One can, in particular, notice that depending on the signs of '
which is more active. All these results are consistent with the view that multimolecular networks are highly organized structures that can spontaneously generate this organization. One can, in particular, notice that depending on the signs of ' 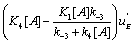 and K 4[A] − K1 [A] then states EA or EB predominate. If state EA predominates this means that the ‘
and K 4[A] − K1 [A] then states EA or EB predominate. If state EA predominates this means that the ‘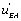 circuit’ predominates as well whereas in the second case it is the
circuit’ predominates as well whereas in the second case it is the 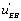 flow’ which is more active. These results are consistent with the view that the “ u' circuits” are highly organized structures that can generate a novel organization. On could feel that the system “chooses” between two different circuits, namely the
flow’ which is more active. These results are consistent with the view that the “ u' circuits” are highly organized structures that can generate a novel organization. On could feel that the system “chooses” between two different circuits, namely the 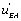 flow ’ and the ‘
flow ’ and the ‘ 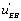 flow’. In fact, it is not a choice but the random interaction of the system with
flow’. In fact, it is not a choice but the random interaction of the system with 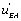 or with
or with 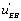 that makes the decision.
that makes the decision.