
Journal of Clinical Trials
Open Access
ISSN: 2167-0870

ISSN: 2167-0870
Research Article - (2021)Volume 11, Issue 5
Background: Several reports have raised safety concerns regarding the use of probiotics. To address these concerns, this study examined the relative abundance (proportion of the microbiome made up of a particular taxa) and normalized read counts (number of times a particular microbe was identified) of Bifidobacteria in the gut microbiome of healthy subjects participating in an ongoing study on the microbiome. Bifidobacteria is a critically important constituent of the human microbiome and plays roles in digestion, gut immunity, and cancer prevention.
Methods: Fecal samples were analyzed using next-generation sequencing to evaluate composition and relative abundance of bacterial phyla through species level in each subject`s microbiome. The primary outcomes of this subgroup analysis were relative abundance and normalized read count of genus Bifidobacteria in subjects who took unregulated probiotics, regulated probiotics, or no probiotics.
Results: The relative abundance and normalized read count of Bifidobacteria were significantly lower in the microbiome of subjects who took unregulated probiotics (n=15) than in the microbiomes of both those who took regulated probiotics (n=12, P=0.0002) and no probiotics (n=13, P=0.0483) (0.18 vs. 9.59 vs. 5.66 relative abundance).
Discussion: Subjects taking unregulated probiotics had a significantly lower relative abundance of Bifidobacteria, which could potentially have a detrimental impact on health. Next-generation sequencing could be a useful tool to guide decisions on the appropriate use of probiotics based on dysbiosis.
Bifidobacterium; Gut microbiome; Probiotics; Whole genome sequencing; Shotgun sequencing
Human host-bacterial symbiosis has been extensively investigated, particularly in the last two decades. Bacteria reside in all parts of our body, with the largest reservoir being the gastrointestinal tract, collectively referred to as the gut microbiome [1]. There is mounting evidence of the critical role of the gut microbiome in human health and disease, which has led to a soaring demand for probiotics and supplements [2]. In 2016, the global probiotics market was valued at approximately $36.6 billion USD, with an anticipated value of $64 billion USD by 2023 [3]. The World Health Organization (WHO) has defined probiotics as “live microorganisms which when administered in adequate amounts confer a health benefit on the host” [4]. Probiotics provide health benefits by modulating gut microbiota. And although some probiotic bacteria are “generally recognized as safe,” several recent reports have raised safety concerns over their use [5]. Concerns raised include risk of systemic infection, metabolic disruption, and immune system overstimulation as well as potential horizontal gene transfer between bacteria (facilitating antibiotic resistance). Cases of bacterial translocation leading to sepsis have been reported following probiotic administration [5].
Study design and participant selection
This was a prospective, longitudinal descriptive study that used shotgun NGS to characterize the gut bacterial microbiome. This study conducted at ProgenaBiome commenced in April 2019 and is currently ongoing. The protocol was approved by Ethical & Independent Review Board and is being conducted in accordance with the applicable requirements outlined in the United States Food and Drug Administration (FDA) Code of Federal Regulations (CFR) Title 21 and International Conference on Harmonisation (ICH) E6 Good Clinical Practice guidelines. All participants signed written informed consent.
During their regular medical visit, men and women of any age who desired to know the composition of their gut microbiome were enrolled in the study. In this study, both healthy volunteers, as well as patients with 34 pre-specified health conditions (Table 1) under examination were included. For the subset analysis, only data from healthy participants were included. Exclusion criteria included history of recent antibiotic use, bariatric surgery and total colectomy with ileorectal anastomosis, proctocolectomy, postoperative stoma, ostomy, ileoanal anastomosis, total parenteral nutrition, or participation in an experimental drug investigation within 12 weeks of our study commencement.
| Organism | P-value | Description of difference |
|---|---|---|
| Bifidobacteria | 0.0001 | Lowest in unregulated group |
| Facaelibacterium | 0.01747 | Lowest in unregulated group |
| Bactroides | 0.391733 | Not significant |
| Clostridium | 0.897584 | Not significant |
| Ruminococcus | 0.003704 | Lowest in unregulated group |
| Eubacterium | 0.027509 | Lowest in unregulated group |
| Alistipes | 0.019949 | Lowest in regulated group, highest in unregulated group |
| Blautia | 0.727962 | Not significant |
| Dorea | 0.911675 | Not significant |
| Coprococcus | 0.677123 | Not significant |
| Roseburia | 0.701591 | Not significant |
| Subdoligranulum | 0.491463 | Not significant |
| Oscillobacter | 0.662898 | Not significant |
| Akkermansia | 0.143468 | Not significant |
| Prevotella | 0.694261 | Not significant |
| Parabacteroides | 0.024904 | Highest in unregulated group |
| Collinsella | 0.528406 | Not significant |
Table 1: Statistical findings for 17 most prevalent genera found in the microbiome of subjects. The p-value for Bifidobacteria is at least one order of magnitude smaller than the other significant genera.
Subjects were selected for this subgroup analysis if they had no known medical issues and had used a single probiotic supplement for at least six weeks prior to sample collection hereafter termed “healthy.”
DNA extraction and sequencing
Fecal samples were collected using either a Zymo Research DNA/RNA Shield Fecal Collection Tube (Zymo Research, Freiburg, Germany) or an OMNIgene Gut Collection Vial (DNA Genotek, Ottawa, CA). Following fecal sample collection, individual subject DNA was extracted and purified using the Qiagen PowerFecal Pro DNA extraction kit (Qiagen, Hilden, Germany). The isolated DNA was then quantified with the Quantus Fluorometer with the QuantFluor ONE dsDNA kit (Promega, Madison, WI, USA). After DNA quantification, the DNA was normalized, and sample libraries were prepared utilizing shotgun methodology with Illumina’s Nextera Flex kit. Sample libraries were then normalized and pooled for sequencing on Illumina’s NextSeq 550 System. Following completion of the NextSeq run, the raw.bcl data were streamed to Illumina’s BaseSpace cloud for conversion to FASTQ files. The FASTQ files were then pushed through the bioinformatics metagenomics pipeline, with subject specific readouts profiling each subject’s unique microbiome.
Study outcomes
All subjects whose gut microbiome was sequenced for this study were volunteers for the larger study on the characterization of the gut microbiome. This subset was selected based on overall health and lack of gastrointestinal symptoms. The primary outcome of this subgroup analysis was Bifidobacterium relative abundance in subjects who used unregulated probiotics, regulated probiotics, or no probiotics. “Unregulated” probiotics were defined as over-the-counter branded pills that are not regulated by the FDA. Probiotics were classified as “Regulated” are FDA approved.
Analysis
Sequencing results were examined for both relative abundance and normalized read counts, both generated by the bioinformatics pipeline. Relative abundance indicates what percentage of the sample is made up of particular taxa of microbes. Normalized read count indicates how many times a particular microbe was identified in a sample, using calculations to correct for the difference in genome size between organisms. Figure 1 was generated using Bifidobacteria relative abundance values from the bioinformatics pipeline. Figure 1 was generated by identifying the 17 most prevalent genera across the groups.
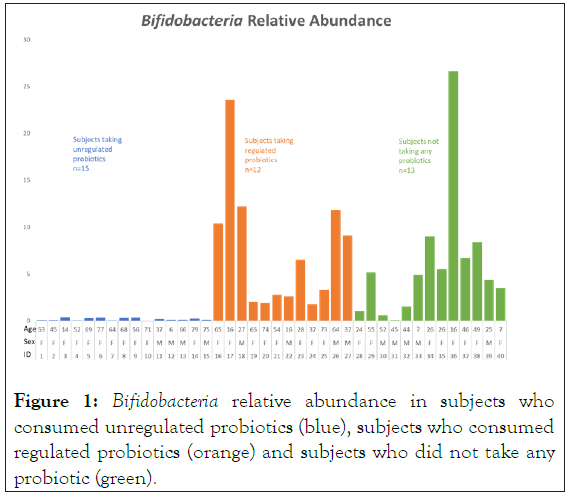
Figure 1: Bifidobacteria relative abundance in subjects who consumed unregulated probiotics (blue), subjects who consumed regulated probiotics (orange) and subjects who did not take any probiotic (green).
Statistical analysis
Continuous variables were presented as mean with Standard Deviation (SD) or median (Interquartile Range [IQR]). ANOVA was performed to identify the difference between the groups. Categorical variables were reported as frequencies and percentages and compared using Pearson χ2 or Fisher's exact test as applicable. A value of P<0.05 was considered statistically significant. The data were analyzed by SPSS version 11.5 for Windows (SPSS Inc., Chicago, IL, USA). The subjects in this analysis comprised the 40 for whom details on the type of probiotics used was available.
Of the 40 subjects included in this study, 15 subjects used unregulated probiotics; 12 subjects used regulated probiotics; and 13, used none. The mean age of the subjects was 46.8 years, and a majority (27/40, [67.5%]) were women. All were considered healthy, with the primary difference being the probiotics taken or not taken, and the relative abundance of Bifidobacteria in their microbiome. While subject age, sex, diet, and blood type were recorded, no correlations were noted between the relative abundance of Bifidobacteria and these variables.
Of the remaining 16 most prevalent genera, only Faecalibacterium, Ruminococcus, Eubacterium, Alistipes, and Parabacteroides were found to be statistically significant (Table 1).
The significance of Bifidobacteria was at least one order of magnitude greater than the other significant genera. As such that became the focus of this study. Shannon Diversity Index was not found to differ significantly between groups.
Figure 1 illustrates the relative abundance of Bifidobacteria for each subject by probiotic group. Subjects taking unregulated probiotics had a lower mean relative abundance of Bifidobacteria in their microbiome than both subjects taking regulated probiotics and subjects who did not take probiotics. The mean Bifidobacteria relative abundance was 0.1[0.04-0.4], 4.9 [0.1-26.6] and 4.9 [1.8-23.57] in the unregulated probiotics, no probiotics, and regulated probiotics groups, respectively (p<0.0001).
Recently, several researchers raised safety concerns regarding the use of probiotics [5,6], particularly in at risk populations, including neonates and those with clinical conditions such as malignancies, autoimmune conditions, diabetes mellitus, and post organ transplantation status. Some probiotic strains may adversely affect those with compromised immunity and potentially facilitate life threatening infections including pneumonia, endocarditis, and sepsis [6,7].
There is a substantial body of “evidence” advocating the benefits of probiotic supplements. However, the quality of evidence as well as the quality of the production of probiotic supplements is matters of concern. In a systematic review of the effects of Lactobacillus casei or Bifidobacterium lactis on a healthy population, 16 studies were evaluated to assess health benefit claims [8]. Of 47 studies listed on the corporate website, all were funded by the company and only 12 overlapped with those meeting inclusion criteria for the systematic review. Only seven of the 12 demonstrated some positive effect following consumption of probiotic enriched milk products. And none of these seven actually demonstrated a clinical benefit to use of the product; rather, they extrapolated from laboratory values. The three pertaining to L. casei received a recommendation grade of D for relying primarily on circumstantial evidence and opinion. In contrast, the four pertaining to B. lactis were of sufficient scientific rigor to merit a recommendation grade of [9,10]. These results suggest that there may be inadequate scientific evidence to support the health claims made for many probiotic products.
The quality of probiotics is also of concern, largely due to the absence of supplement regulation. In a national survey of commercial probiotic food supplements in Italy, of 25 samples that claimed B. bifidum was present, none contained the live bacterium and the non-viable bacterium was only sporadically present [11]. More importantly, some of these supplements contained toxin producing species of bacteria including Bacillus cereus. Another study that evaluated 16 probiotic products found the contents of only one brand to match its label claims. And even within that particular brand, inter and intra-lot variation was noted [12]. In many cases the products contained little or no viable bacteria. A third study involving 26 probiotic products found instances of misidentification of the bacteria at both the species (27 incorrectly identified) and genus levels (19 incorrectly identified) [13,14]. This misidentification is clinically significant. As quoted by Rijkers et al., “Stig Bengmark has made this very clear in his statement that the (genetic) difference between one probiotic bacterium and the other is larger than the difference between a man and a goldfish.” This is illustrated by a study which compared the activity of two different Lactobacillus strains (Lactobacillus salivarius CECT5713 and Lactobacillus fermentum CECT5716) in mice. Despite the similarities between the strains, L. fermentum was found to be immunostimulatory, whereas L. salivarius was found to have an anti-inflammatory response [15]. When dietary supplement production facilities were inspected in 2019, over half (305/598) were issued Form 483 for violations of manufacturing standards and safety. The most commonly cited violation categoryaccounting for 25% of violations was failure to adequately describe the final product, including identity, purity, and strength. Other violations included an absence of written quality control procedures, failure to follow written procedures, and incomplete batch records [16]. Poor quality control was identified as the main factor leading to non-compliance. Given the staggering lack of quality control checks in the probiotic industry, choosing an appropriate probiotic supplement is challenging.
Because 1) probiotics can adversely modify gut flora, 2) clinical studies on their effects are of questionable quality, and 3) probiotics lack clear endorsement or approval by regulatory authorities or agencies, choosing a suitable probiotic poses a considerable challenge. Determining the composition of an individual’s microbiome prior to the start of therapy, would allow targeted rather than “empiric” therapy. Such a personalized approach could serve as a safety measure for preventing possible infection and pathologic dysbiosis, which could mitigate associated morbidity and mortality [17]. Next- Generation Sequencing (NGS) of the gut microbiome could become an important tool for identifying gut dysbiosis.
In this study we use next-generation sequencing of the gut microbiome to determine composition and relative abundance in association with the clinical characteristics of patients, following probiotic administration. We describe the results of a subgroup analysis performed to identify changes in microbiota in terms of fecal Bifidobacterium counts in patients taking probiotics dichotomized on regulation or lack thereof compared to those who did not take probiotics.
Citation: Jordan D, Andreas P, Brad B, Sabine H (2021) Impact of Probiotics on Gut Microbiome Bifidobacterium Relative Abundance: First Do No Harm. J Clin Trials. 11:473.
Received: 28-Jul-2021 Accepted: 11-Aug-2021 Published: 18-Aug-2021 , DOI: 10.35248/2167-0870.21.11.473
Copyright: © 2021 Jordan D, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.