Fungal Genomics & Biology
Open Access
ISSN: 2165-8056
ISSN: 2165-8056
Mini Review - (2022)Volume 12, Issue 3
During autophagy in Saccharomyces cerevisiae, cytosolic components are engulfed by double-membrane compartments called autophagosomes, transported to the vacuole, and degraded there. The cytoplasm-to-vacuole targeting (Cvt) pathway delivers the vacuolar hydrolase aminopeptidase I (Ape1) through double-membrane compartments termed Cvt vesicles. These pathways share common mechanisms including the degradation of their compartments. The outer membranes of autophagosomes and Cvt vesicles fuse with the vacuole membrane and inner membranebound structures and, autophagic bodies and Cvt bodies, are released into the vacuole lumen. The membranes of autophagic bodies and Cvt bodies are degraded by a vacuolar lipase, Atg15, to allow other vacuolar hydrolases to access their cargoes for degradation. Atg15 is the only vacuolar lipase in S. cerevisiae and has a transmembrane domain at the N-terminus and a lipase domain at the C-terminus. Recently, our group has clarified the roles of these two domains in vivo. Here we review how Atg15 functions for degradation of the membranes of autophagic bodies and Cvt bodies in the vacuole.
Atg15; Autophagic body; Autophagy; Cvt pathway; Lipase; Saccharomyces cerevisiae
In Saccharomyces cerevisiae, autophagy is induced by starvation or rapamycin treatment. Upon induction of autophagy, autophagy-related (Atg) proteins are recruited to the vacuolar membrane to generate a double membrane–bound organelle, an autophagosome, with engulfing cytoplasmic components [1,2]. The outer membrane of an autophagosome fuses with the vacuolar membrane, and the inner membrane compartment of the autophagosome, the autophagic body, is released into the vacuolar lumen. S. cerevisiae undergoes a specific type of selective autophagy, called the cytoplasm-to-vacuole targeting (Cvt) pathway, which delivers the vacuolar hydrolase aminopeptidase I (Ape1) under nutrient-rich conditions [3-5]. In this pathway, precursor Ape1 (prApe1) assembles into the Ape1 droplet [6]. The Ape1 droplet is engulfed by a double-membrane vesicle, the Cvt vesicle, and transported to the vacuolar lumen. Similar to an autophagosome during starvation, the outer membrane of the Cvt vesicle fuses with the vacuole membrane and the vesicle derived from the inner membrane compartment, the Cvt body, is released into the vacuolar lumen. Membranes of autophagic bodies and Cvt bodies should be disintegrated to allow vacuolar hydrolase to access their cargoes for degradation. The most remarkable difference between an autophagic body and a Cvt body is their size; an autophagosome is 300–900 nm in diameter, whereas a Cvt vesicle is 140–160 nm in diameter [7,8]. Atg15/ Aut5/Cvt17, a glycosylated integral membrane protein that is essential for the disintegration of autophagic bodies and Cvt bodies, is the only vacuolar lipase in S. cerevisiae [9,10]. Atg15 has a transmembrane domain (TMD) at its N-terminus and a lipase domain at its C-terminus [9]. The serine residue at position 332 in its C-terminal region is supposed to be the active center of Atg15 as a lipase [9-11]. On the other hand, immunofluorescence microscopy demonstrates that Atg15 localizes to the endoplasmic reticulum (ER) and is transported to the vacuole via the multivesicular body (MVB) pathway [9]. It remains unknown whether the N-terminal TMD plays a role in the Atg15 activity and whether the C-terminal lipase domain actually functions in the vacuolar lumen. Here we review the recent research by our group that analyzes the functions of these two domains of Atg15 [12].
In previous research, transport of Atg15 has been shown in fixed cells [9,13]. In our study, C-terminal 2 × GFP-tagged Atg15 (Atg15-GFP) was constructed to visualize the dynamics of Atg15. Atg15-GFP localized to the vacuolar lumen and ER in wild-type cells [12]. Furthermore, Atg15 was delivered to the vacuole via the MVB pathway. Without Pep4, an aspartyl protease that plays a central role in the maturation of vacuolar hydrolases, Atg15-GFP was visualized as small dots moving around inside the vacuolar lumen. This is consistent with a previous report that Atg15- localizing vesicles derived from the MVB pathway accumulate in the vacuolar lumen in pep4Δ cells [9]. Atg15 degrades not only autophagic bodies and Cvt bodies, but also MVB vesicles [13]. These facts demonstrate that Atg15 functions inside the vacuole after it is transported into the organelle.
To examine the role of N-terminal TMD of Atg15 (Atg15TMD), we constructed GFP-fused Atg15TMD, 2–36 amino acid residues of Atg15. GFP-Atg15TMD was transported to the vacuolar lumen via the MVB pathway in the same way as the full-length Atg15-GFP protein. This showed that Atg15TMD (amino acid residues 2–36) is sufficient for Atg15 delivery to the vacuole via the MVB pathway. Next, to investigate whether the N-terminal TMD is required for the function of Atg15, we altered the transport pathway of Atg15 by replacing its original N-terminal domain with the N-terminal domain of Pho8, which is transported to the vacuole via the Pho8 pathway [14]. We fused GFP to the N-terminus of Pho8, Pho8TMD, which serves as the sorting signal sequence of the Pho8 pathway, and the C-terminal region of Atg15 (amino acid residues 37–520, including the lipase domain) to generate GFP-Pho8TMD-Atg15C (Figure 1). This GFP-Pho8TMD-Atg15C was successfully transported to the vacuolar membrane and was functional for the degradation of autophagic bodies and Cvt bodies. This result demonstrated that the delivery of Atg15 via the MVB pathway is not essential. Rather, transport of the C-terminal region to the vacuole is crucial for its activity. It is probable that the main function of the N-terminal TMD is for the transport of the C-terminal region to the vacuole. Previously, Teter et al. has proposed that several highly conserved residues constitute the catalytic triad: S332, D387 or D421, and H435 [10]. We showed that residue D387, not D421, is important for the activity of Atg15, suggesting that D387 consists of the catalytic triad. Moreover, using C-terminal truncation mutants, we showed that 50–466 residues are sufficient for its activity. The W466 residue, which is important for the activity of Atg15, is highly conserved at the extreme C-terminal region among Atg15 orthologs. An alanine mutation at this residue showed a defect in the degradation of autophagic bodies, not Cvt bodies. These findings indicate that W466 is critical for the degradation of autophagic bodies, which are relatively larger compartments than Cvt bodies.
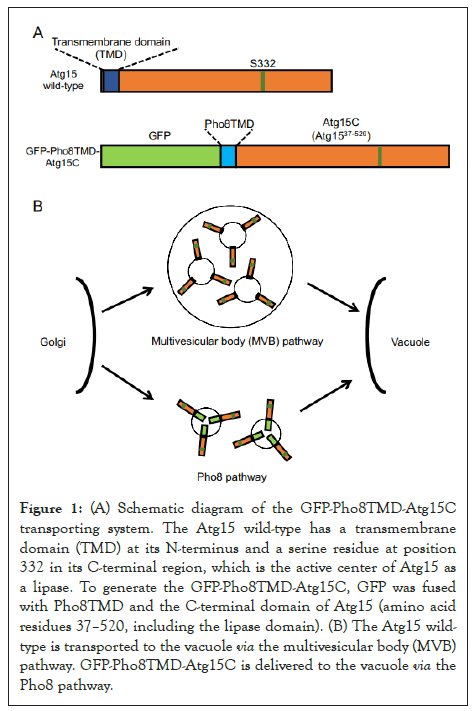
Figure 1: (A) Schematic diagram of the GFP-Pho8TMD-Atg15C transporting system. The Atg15 wild-type has a transmembrane domain (TMD) at its N-terminus and a serine residue at position 332 in its C-terminal region, which is the active center of Atg15 as a lipase. To generate the GFP-Pho8TMD-Atg15C, GFP was fused with Pho8TMD and the C-terminal domain of Atg15 (amino acid residues 37–520, including the lipase domain). (B) The Atg15 wildtype is transported to the vacuole via the multivesicular body (MVB) pathway. GFP-Pho8TMD-Atg15C is delivered to the vacuole via the Pho8 pathway.
In this study, we demonstrated that Atg15 has two functional domains; the N-terminal TMD plays a role in the transport of Atg15 to the vacuole via the MVB pathway and the C-terminal catalytic domain is essential for membrane degradation inside the vacuole. However, the MVB pathway is not required for the degradation of autophagic bodies and the C-terminal domain inside the vacuole is sufficient for its function. Considering these facts, it is possible to hypothesize that Atg15 is inactive before it reaches the vacuole and is activated once inside. Pep4 is an aspartyl endopeptidase that cleaves Prb1 and Pho8 in order to activate them [15]. Likewise, Pep4 might activate Atg15 by cleaving it. We demonstrated that Atg15-localizing MVB vesicles accumulate inside the vacuolar lumen in pep4Δ cells. Given the fact that Atg15 degrades MVB vesicles [13], this may be because Atg15 in this mutant is not activated without Pep4. If this hypothesis is true, the truncation mutant of Atg15, which has an essential minimal region for its function, can bypass the activity of Pep4. However, the minimal truncation mutant of Atg15 (residues 50– 466: GFP-Pho8TMD-Atg15ΔN49ΔC54), which we determined in this study, did not degrade autophagic bodies without Pep4.
There are two possibilities for this defect. First, the GFPPho8TMD moiety and the Atg15ΔN49ΔC54 moiety must be cleaved by Pep4. When GFP-Pho8TMD-Atg15ΔN49ΔC54 is expressed in cells, the GFP-Pho8TMD moiety can be detected by immunoblot analysis, suggesting that the GFP-Pho8TMD moiety is cleaved from GFP-Pho8TMD-Atg15ΔN49ΔC54 inside the vacuole. Atg15ΔN49ΔC54 may then play a role in the degradation of autophagic bodies, although it is still possible that this portion undergoes further cleavage by Pep4. Since the N-terminal truncation mutant of Atg15 [(Atg15ΔN63 (Atg1564– 520), Atg15ΔN70 (Atg1571–520), and Atg15ΔN90 (Atg1591–520)] shows abnormal localization and cannot reach the vacuole, even if fusing with Pho8TMD moiety, we cannot determine whether this region plays a role in its activity. Another challenge to analyzing the cleaved Atg15 portion is that we have never detected the Atg15ΔN49ΔC54 moiety in GFP-Pho8TMDAtg15ΔN49ΔC54– expressing cells by immunoblot analysis (unpublished observations). Previous studies have indicated that full-length Atg15 is a short-lived protein with a half-life of less than 1 h [9,10]. This might be one of the reasons that processed Atg15 could not be detected. The second hypothesis is that Pep4 has an additional role in the degradation of autophagic bodies, other than the cleavage of Atg15. Further experiments are required to elucidate this possibility.
Atg15 disintegrates vesicles from the MVB, Cvt, and autophagic pathways [13]. We found that the W466 residue of Atg15 is required for the degradation of autophagic bodies, whereas not for Cvt bodies. One of the differences between these vesicles is their size, in other words, the curvature of the membrane. The average sizes of MVB, Cvt, and autophagic vesicles are ~25 nm, 140–160 nm, and 300–900 nm, respectively [7,8,16]. It might be possible that Atg15 normally changes its conformation to recognize various membrane curvatures. However, when W466 is mutated, Atg15 may lose flexible conformation and cannot interact with high curvature membranes. Further analysis of the degradation activity of various sized vesicles by Atg15 in vitro is needed to examine this hypothesis. It is also possible that Atg15 recognizes specific molecule(s) on the surface of autophagic bodies and Cvt bodies and W466 is important for that system.
One fundamental question remains. If Atg15 can disintegrate biological membranes nonspecifically, why does it not degrade the vacuolar membrane? There are two possible answers to this question. One is that Atg15 selectively degrades intravacuolar vesicles rather than the vacuolar membrane. Atg15 might recognize a specific lipid residing on the membrane of intravacuolar vesicles as its substrate. Otherwise, Atg15 might be responsible for the positive curvature of intravacuolar vesicles. A second possibility is that the vacuolar membrane is actually disintegrated by Atg15, and is immediately repaired by the membrane reconstitution system.
To analyze these hypotheses, purification of active Atg15 is required. Since active Atg15 is present at least inside the vacuole, it should be a straightforward process to purify Atg15 from the vacuole. Autophagic bodies are a suitable substrate to measure the activity of purified Atg15. Isolation of subvacuolar vesicles, which contain prApe1, has already been achieved [17], and this method should also apply to the isolation of autophagic bodies and Cvt bodies. We have established this system using N-terminally mRFP-fused prApe1 to monitor the degradation of autophagic bodies in vivo [12]. In vitro assay of mRFP-prApe1 could also be a useful marker to monitor the degradation of autophagic bodies and Cvt bodies.
In summary, we have shown that Atg15 has two distinct functional domains. The N-terminal transmembrane domain is required for the transport of Atg15 to the vacuole via the MVB pathway. On the other hand, the C-terminal lipase domain is responsible for its enzymatic activity inside the vacuole.
For future research, it will be important to develop an in vitro monitoring system for the activity of Atg15 to analyze the mechanisms of degradation of autophagic bodies and Cvt bodies. Our group is currently attempting to purify active Atg15 together with its substrate. After purification of active Atg15, we will be able to monitor the degradation activity of Atg15 for various substrates, for example, autophagic bodies, Cvt bodies, MVB vesicles, and artificial liposomes. Using this system, it will be possible to analyze the degradation activity of Atg15 for substrates with various sizes and membrane compositions. Moreover, purified Atg15 can also be used for crystal structure analysis. These investigations should provide important information about the conformation of Atg15 during interaction with membranes. Through these analyses, we will be able to understand how Atg15 recognizes and degrades various biological membranes inside the vacuole.
This work was supported by a grant from the Naito Foundation (to KS) and by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (21K15093 to EH and 20H05313, 21K19205, and 22H02569 to KS). This work was supported by CREST, the Japan Science and Technology Agency, Japan (JP201032912 to KS).
Citation: Hirata E, Suzuki K (2022) How do Two Domains of Atg15 Work for Autophagosomal Membrane Degradation? Fungal Genom Biol. 12:188.
Received: 23-Apr-2022, Manuscript No. FGB-22-17118; Editor assigned: 26-Apr-2022, Pre QC No. FGB-22-17118 (PQ); Reviewed: 12-May-2022, QC No. FGB-22-17118; Revised: 18-May-2022, Manuscript No. FGB-22-17118 (R); Published: 25-May-2022 , DOI: 10.35841/2165-8056.22.12.188
Copyright: © 2022 Hirata E, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.