Fungal Genomics & Biology
Open Access
ISSN: 2165-8056
ISSN: 2165-8056
Research - (2023)Volume 13, Issue 4
Laccase exists widely in plants and fungi. It is a copper-containing polyphenol oxidase that can degrade lignin, oxidate and phenolic substances, inhibit heterophytes, promote fruiting body formation and improve the quality of mushrooms. In this study, 18 laccase genes were identified from the whole genome of a white strain of Hypsizygus marmoreus (HM62). These genes were mainly distributed on chromosomes 1, 2, 3, 4, 6, 9 and 10. Additionally, 9 genes were found to be clustered linearly on chromosome 6, indicating gene duplication. Multiple sequence alignment and protein similarity comparison revealed the presence of polymorphism among these laccase genes. Phylogenetic analysis showed that the laccase gene family of H. marmoreus was classified into four subfamilies. The spatiotemporal expression analysis of the laccase gene family showed that HmLac09 and HmLac10 were highly expressed in different periods and might be involved in lignin degradation and fruit body formation, respectively. The expression levels of HmLac02, HmLac05, HmLac08 and HmLac17 genes in gray or gray and white heterozygous strains were higher than those in white strains, which might be related to the difference in lignin decomposition in gray strains and one of the factors leading to different growth rates. The present study investigated the characterization of the H. marmoreus laccase gene family, extending our understanding of laccase mediated fruiting body development and growth rate mechanisms in this fungus.
Hypsizygus marmoreus; Laccase; Genome identification; HmLacs gene
Laccase is a polyphenol oxidase with copper ions. It is involved in the degradation of lignin together with lignin peroxidase, manganese peroxidase and multifunctional peroxidase and is widely present in plants, insects, fungi and bacteria [1,2]. The laccase molecule is composed of a single polypeptide, a copper ion active center and a sugar ligand. Those from different sources vary in degrees of glycation that use the unique redox ability of copper ions to carry out one-electron oxidation of reducing substrates and reducing oxygen to water [3]. According to the nature of magnetism and spectroscopy, laccase is composed of three conservative copper ion structural domains. The active center of the copper ions is divided into three categories; Type I Copper ion (T1-Cu) or blue type copper (T2), Type II Copper ion (T2-Cu) or blue hydrated type copper and two Types III Copper ions (T3-Cu) or coupling double karyotypes of copper [4,5].
However, all laccase structures do contain all three types of copper ions. Typically, copper ion has four non-vacancy conserved motifs (L1–L4), which are the marker sequences of laccase from another polyphenol oxidase. These sequences include 10 histidines and 1 cysteine. These amino acid residues combine with the three copper ions of laccase to form ligands that effectuate the physiological roles of laccase [6-8]. Laccase needs various functions to adapt to different growth environments; thus, they gradually differentiate into varied homologous genes with different functions (combined sequence and structure analysis of the fungal family). In the evolution of laccase protein, some related functional amino acid residues rarely mutated and became a conserved part, which was used as the identification tag of the gene (the structure and function of fungal laccases).
In addition, laccase genes were involved in lignin degradation, vegetative growth, fruiting body formation and pigmentation during the growth of edible fungi [9]. Laccase gene families have been reported in edible fungi, such as Coprinopsis cinerea, Auricularia auricula, Flammulina velutipes, Volvariella volvacea and Pleurotus ostreatus. To date, 17 gene families in Coprinopsis cinerea, the largest basidiomycete’s laccase gene family, have been identified [10-14]. Several studies have assessed the molecular and functional aspects of this family. The laccase gene of H. marmoreus (lcc1), 2336 base pairs (bp) in length containing 13 introns and 14 exons, was cloned and the phylogenetic tree showed homology with the laccase gene of Flammulina velutipes. Interestingly, the laccase activity of the recombinant strain was higher than that of the control, the growth rate of mycelia was significantly increased, the primordia formation was 3-5 days early and the fruiting body maturity was 5 days higher, indicating that the laccase gene could promote the growth of mycelia and the development of the fruiting body [15]. A previous study showed that the activities of laccase and β-glucosidase in the primordia stage were significantly higher than those in other states, which might be related to the formation of primordia and promote the early transfer reproductive growth of H. marmoreus [16]. Kojic acid is an inhibitor of laccase; a study showed that laccase activity was downregulated by kojic acid during mycelia recovery and color transformation but significantly upregulated during the primary stage, further indicating that laccase is closely related to the fruiting body development of H. marmoreus [17].
Hitherto, only a few studies have evaluated the laccase genes due to the lack of a laccase genome and systematic identification, induction and functional analysis of the laccase gene family. Therefore, in the present study:
• The laccase gene family was systematically identified and its structure and chromosomal location were analyzed at the chromosome level of the white strain genome.
• Intraspecific and interspecific evolution of the laccase gene family in H. Marmoreus was assessed.
• The spatiotemporal expression of the laccase gene in different tissues and mycelia of H. Marmoreus in different periods was analyzed.
The materials
Three transcriptomic experiments were carried out to analyze the spatiotemporal expression of laccase genes. In the first experiment, three stages of mycelia post-ripening stage (opening and tiling bacteria), a color turning stage (gray strain turning color, white strain not turning color) and primordia formation were selected during the growth of the H. marmoreus grey strain (Hm61) and white strain (Hm88). In the second experiment, the lid epidermal tissue samples were taken from gray (Hm61), white (Hm88) and their hybrid progeny (HMZ5) strains. In the third experiment, mononuclear mycelium Hm61_G6, Hm88_W2 and their hybrid HMZ5 mycelium were respectively taken. There were three biological replicates per sample in each experiment. The cultivation bag matrix (mass ratio) values were wood 78%, bran 21%, lime powder 0.5%, gypsum powder 0.5% and water content 65%.
Sequence retrieval
The white strain HM62-W was the reference genome, which was completed by Genome and Biotechnology Research Center of Strait Joint Research Institute of Fujian Agriculture and Forestry University (NCBI Accession no. JABWDO000000000). The reference genome of the grey strain Haemi51987-8 was published in 2018. Combined with High‐Throughput Chromosome Conformation Capture technique (Hi-C) sequencing data, 278 overlapping groups of Haemi51987-8 were interrupted for remounting and gene prediction, resulting in a high-quality genome map named HM01_Gray. Macrolepiota albuminosa data from Ensembl Fungi database, while the data of Wolfiporia extensa and Fistulina hepatica were obtained from NCBI database.
Identification of members of laccase gene family of H. marmoreus
The identification procedures of laccase gene family members of Laccase of H. marmoreus were as follows:
• Using NCBI GenBank (LCC1-LCC17: Bk004111-bk004127) and the amino acid residues of 17 non-allelic laccase genes from Coprinopsis cinerea were used as seed sequences. With the help of local Blast software, the sequences with E value less than or equal to 1e-10 were used as candidate bases [18,19].
• Candidate sequences were compared back to Swissport database and the sequence with the highest consistency was reserved laccase gene.
• Through multiple sequence alignment, the genes without laccase marker sequences were deleted.
• Used the Batch Command (CD)-search function of Consular Consolidated Database (CDD) database to predict the domain of candidate genes and deleted the genes with iron oxidase domain.
To avoid the loss of possible laccase gene family members due to incomplete domains, the Self-Monitoring, Analysis and Reporting Technology (SMART) database was used to verify the existence of three conserved domains. The candidate genes with laccase conserved domain were selected as members of the HmLacs family. The identification method of laccase gene family members of 54 strains of H. marmoreus, Macrolepiota albuminosa, Wolfiporia extensa, Fistulina hepatica and other edible fungi was the same as above.
Use SignalP 5.0 Server and SecretomeP 2.0 Server to predict HmLacs family typical signal peptide and atypical signal peptide. GOR4 was used for secondary structure prediction of HmLac proteins and SWISS-MODEL online tools were used for 3D protein structure prediction.
Gene structure and conserved motif analysis of HmLacs
MEME website tool was used for HmLacs family Motif (Motif) identification and analysis of the Motif width was set to 6-200 residue, the biggest base sequence number for 25, repeat any number of times using Python scripts to obtain chromosome positions and exon-intron numbers using TB tools for the phylogenetic tree, gene structure and conservative Motif distribution visualization [20].
Chromosomal localization and three-dimensional structure of all HmLacs
MCScanX software was used to analyze the collinearity and gene duplication events. The E value of blast was less than or equal to 1e-10 and the other parameters were default parameters [21]. For tandem repetition file output by MCScanX, further manual identification was carried out and the identification criteria were as follows:
• The ratio of shorter sequence length to longer sequence length was large at 70%.
• The similarity of the two amino acid sequences is more than 70%.
• The two genes were in 100 KB fragment [22].
• Tb tools was used to display chromosome location [23].
Multiple sequence alignment, phylogenetic analysis and classification of H. marmoreus laccases
The intraspecific (white and gray strain) and interspecific (Coprinus cinereus, Pleurotus ostreatus, Flammulina velutipes, Lentinula edodes, Volvariella volvacea, collybia albuminosa, Wolfiporia extensa, Fistulina hepatica) laccase gene family phylogenetic trees were constructed respectively and the protein sequences of laccase gene in Arabidopsis thaliana were outgroup. Phylogenetic tree construction was carried out with the help of the fin suite manager plug-in [24]:
• Multi-sequence alignment of protein sequences using the normal alignment mode of Mean Time to Failure (MATTF).
• Deletion of vacancies using trim Advanced Technology Attachment (ATA) interface and retention of conserved amino acid residues.
• Modelfinder software selects the best protein evolution model [25].
• Under the model of IQ-tree automatic selection (automatic option in IQ-Tree) [26]. The maximum likelihood phylogenetic TREE was deduced for 20000 ultra-fast guidance and approximate likelihood ratio tests using IQ-tree [27,28].
• For the construction of laccase family sequence of gray strain and white strain of Mushroom, phylogenetic tree was constructed by Bayesian method and MrBayes was used under Wag+I+G model 3.2.6 Software reconstruction of Bayesian phylogenetic tree, sampling every 100 generations, discarding 25% of aging samples, remaining samples tree construction and calculation of posterior probability [29].
Analysis of the expression profiles of HmLacs in H. marmoreus based on RNA-seq
Total RNA was extracted from each sample according to the Kit (E.Z.N.A Plant RNA Kit, Omega, Biotech, Norcross, Ga). Illumina NEBNext® UltraTM RNA Library Prep Kit was used for Library construction and the steps provided by the Kit were followed. Total RNA samples were commissioned to be sequenced using Illumina HiSeqTM 2500 (Illumina Inc, CA, USA) platform by Beijing Nuohe Zhiyuan Bioinformatics Technology Co. LTD. Sequencing depth of each sample was 60X. At the same time, some RNA samples were kept in the -80℃ refrigerator for storage.
The transcriptome data analysis steps are as follows:
• All Illumina data sequenced by RNA-seq were subjected to quality control by FastQC and Trimomatic [30].
• The HISAT2 software was used to compare RNA sequencing read segments with default parameters and Samtools was used to sort alignments according to the order of reference sequences, so as to obtain THE ALIGNMENT results in Binary Alignment and Map (BAM) format [31,32]. Statistics were made according to the alignment results.
• Based on the comparison results, Stringtie software was used to estimate gene expression [33]. In addition, Ballgown software provided by Stringtie were used to extract the number of read segments (read count) and FPKM values of genes located in gene exons after comparison from the results generated by Stringtie [34].
• edgeR read counts was used to convert it into CPM (Counts -Per Million) to filter genes that were not expressed or were all low expressed in all samples [35].
• Differential expression analysis was conducted by Pairwise comparison of each sample using Bioconductor's R language package edgeR: After filtering with CPM values and Normalization with TMM (Mean of M-values), crosstalk significance is checked using statistical methods from edgeR (p values are calculated), the fold change between the two groups was estimated. Visualization of Differentially Expressed Genes (DEG) using R language is identified.
• According to the list of differentially expressed genes, we used R language Bioconductor package Topology of Gene Ontology (TopGO) for enrichment analysis. TopGO estimates the significance of functional enrichment (p-value) based on the hypergeometric distribution into Fisher's exact probability test. In the enrichment analysis results, p-value ≤ 0.05 was taken as the threshold and the functions meeting this condition were defined as significantly enriched functions.
• Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed by R/Bioconductor package according to the list of differentially expressed genes. Enricher function was used to estimate the significance of functional enrichment (p value). With p-value ≤ 0.05 as the threshold, functions meeting this condition were defined as significantly enriched functions.
Experimental validation of HmLacs gene expression levels by qRT-PCR
The expression levels of DEGs were validated by quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR). Primer3Plus and NCBI Primer-BLAST were used to design gene-specific primer pairs. Total RNA of tissues was extracted using Invitrogen Trizol, first-strand complementary DNA (cDNA) was synthesized with StarScriptⅡFirst-strand cDNA Synthesis mix with genomic Deoxyribonucleic Acid DNA (gDNA) Remover for qPCR (A224-02; Genstar). The RT-qPCR was performed with 2x RealStar Green Fast Mixture (Genstar) on Multicolor Real-Time PCR Detection System (Bio-Rad). Reaction parameters for thermal cycling were 95ºC for 2 min, followed by 40 cycles of 95ºC for15 s and 60ºC for 30 s, finally a melting curve (65-95ºC, at the increments of 0.5ºC) performed to confirm the PCR specificity. The expression level of each gene relative to housekeeping genes were calculated using the 2-∆∆Ct with three replicates per sample [4]. Then we made the correlated analyses between qRT-PCR values with Fragments per Kilobase of Transcript per Million (FPKM) values. In this experiment Glyceraldehyde-3-Phosphate Dehydrogenase (GADPH) and β-actin were used as the reference genes.
Identification of genes and characterization of homologous genes encode the laccase proteins in H. marmoreus
A total of 20 laccase genes were identified as candidates in the whole genome by gene family identification and similarity search against the published model fungi of C. cinerea, P. ostreatus, F. velutipes and L. edodes. The comparison between CDD, SMART and Swissport databases and analysis of laccase marker sequences (L1-L4) identified 18 laccase genes in the reference genome hm62-W of the white strain (Figure 1), all these genes had three copper ion conserved domains. The amino acid sequences of 18 laccase genes were consistent with the characteristics of fungal laccase and no deletion or replacement of amino acid residues was detected. Also, the characteristic sequence of fungal laccase was L1-L4, i.e. the binding region of copper ions. At these sites, copper ions T1, T2 and T3 must bind to 10 histidines and 1 cysteine to form functional ligands (Figure 2).
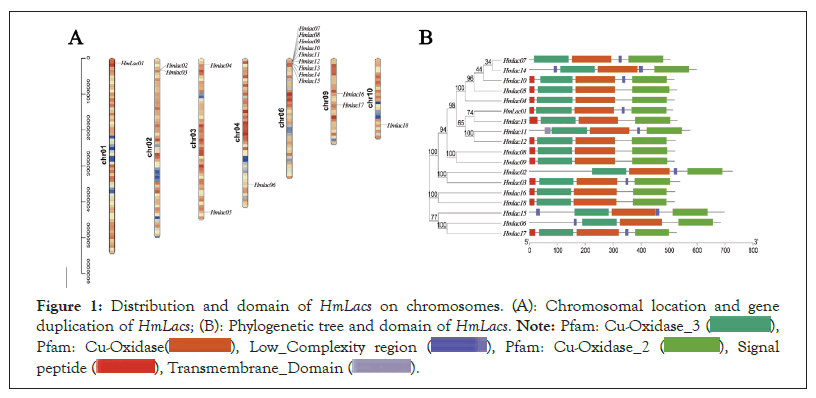
Figure 1: Distribution and domain of HmLacs on chromosomes. (A): Chromosomal location and gene
duplication of HmLacs; (B): Phylogenetic tree and domain of HmLacs. 
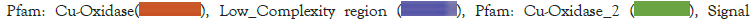

Figure 2: Gene structure and protein similarity comparison. (A): Standard sequence (L1-L4) multiple sequence alignment of H. marmoreus and Motif results; (B): Laccase family gene amino acid sequence identity comparison.
The number of amino acids encoded by the 18 laccase gene proteins in the reference genome of laccase is 504-726 and the molecular weight is 53.41356-80.99129 kDa (Table 1). The Isoelectric Point (IEP) was 4.41-6.52 except for HmLac02, which was 8.45. Compared to most fungal laccase, the IEP was in line with the physicochemical properties of fungal laccase. HmLac02 was identified as a basic protein, which might have other functions of laccase.
| Gene Name | Gene ID | Size (aa) | MW (Da) | pI | Chromosome number | Signal peptide | Intron/exon number |
|---|---|---|---|---|---|---|---|
| HmLac01 | HM01gene000570 | 513 | 55329.78 | 6.2 | 1 | 16-17 | 15/14 |
| HmLac02 | HM01gene018960 | 726 | 80991.29 | 8.54 | 2 | - | 24/24 |
| HmLac03 | HM01gene018970 | 538 | 58712.2 | 5.52 | 2 | 22-23 | 14/14 |
| HmLac04 | HM01gene035380 | 518 | 55068.24 | 4.41 | 3 | 18-19 | 15/17 |
| HmLac05 | HM01gene051280 | 526 | 56023.63 | 4.99 | 3 | 18-20 | 15/20 |
| HmLac06 | HM01gene064720 | 683 | 74349.71 | 6.52 | 4 | - | 22/25 |
| HmLac07 | HM01gene082910 | 504 | 53413.56 | 4.45 | 7 | - | 15/21 |
| HmLac08 | HM01gene082960 | 520 | 56522.73 | 6.07 | 7 | 21-22 | 12/12 |
| HmLac09 | HM01gene082970 | 519 | 56568.31 | 3.4 | 7 | 21-22 | 12/16 |
| HmLac10 | HM01gene083010 | 517 | 54974.59 | 4.82 | 7 | 18-19 | 15/19 |
| HmLac11 | HM01gene083030 | 575 | 61995.57 | 3.41 | 7 | - | 18/18 |
| HmLac12 | HM01gene083050 | 521 | 55900.59 | 6.1 | 7 | 18-19 | 15/17 |
| HmLac13 | HM01gene083150 | 529 | 56965.73 | 5.94 | 7 | 29-30 | 15/21 |
| HmLac14 | HM01gene083210 | 597 | 63908.77 | 5.08 | 7 | - | 18/18 |
| HmLac15 | HM01gene083660 | 697 | 76917.7 | 6.05 | 7 | - | 28/28 |
| HmLac16 | HM01gene117180 | 520 | 57214.98 | 6.34 | 9 | 19-20 | 30/30 |
| HmLac17 | HM01gene118350 | 525 | 56864.6 | 6.56 | 9 | 21-22 | 22/24 |
| HmLac18 | HM01gene128560 | 519 | 56701.19 | 5.98 | 10 | 18-19 | 23/23 |
Table 1: Features of HmLacs genes identified in Hypsizygus marmoreus.
The cleavage location of the laccase gene signal peptide was predicted and the results showed that HmLac01, HmLac03, HmLac04, HmLac05, HmLac08, HmLac09, HmLac10, HmLac12, HmLac13, HmLac16, HmLac17. Twelve genes, including HmLac18, had signaling peptides at the N-terminal about 16–30 Attoampere (Aa) long, identified as secretory proteins. On the other hand, the 6 kinase genes, HmLac2, HmLac6, HmLac7, HmLac11, HmLac14 and HmLac15 were found to lack the typical signal peptides (Table 1). However, in the prediction of subcellular localization, the results indicated that all 18 HmLacs were extracellular proteins. Additionally, the prediction of atypical laccases revealed the presence of atypical signaling peptides. This suggests that not all members of the laccase gene family possess the function of lignin degradation.
Genomic location and duplication events among HmLac genes
In the reference genome, the 18 laccase genes were primarily located on chromosomes 1 (HmLac01), chromosomes 2 (HmLac02 and HmLac03), chromosomes 3 (HmLac04), chromosomes 5 (HmLac05), chromosomes 4 (HmLac06), chromosomes 6 (HmLac07, HmLac08, HmLac09, HmLac10, HmLac11, HmLac12, HmLac13, HmLac14 and HmLac15), chromosome 9 (HmLac16 and HmLac17) and chromosome 10 (HmLac18). Herein, we also found that these 18 laccase genes, except HmLac16 and HmLac17 distributed in the middle of chromosome 9, were mainly distributed at both ends of the chromosome and the chromatin in this region was loose, which needs to be investigated further. Noteworthy observations, the laccase genes HmLac07, 08, 09, 10, 11, 12, 13 and 14 are clustered on chromosome 6. Similarly, HmLac02 and HmLac03 are clustered on chromosome 2 and their amino acid sequences exhibit a high degree of similarity, while the distance between them is less than 10 kb. This indicates that they are tandem duplicated genes and suggests that the laccase gene family underwent gene duplication events during the evolutionary process. Combined with the MCScanX operation and manual correction results, we found that HmLac10 and 14, HmLac11 and HmLac13 were lineal homologous genes. HmLac02, 03, 08, 09, 10, 11 and 12 genes formed the tandem repeats and HmLac01, 04, HmLac05, 06, 13, 14, 15, 16, 17 and 18 may be produced by retro transposition. The 18 HmLac proteins of H. marmoreus are primarily composed of α-helices, β-sheets and random coils. Among them, the highest proportion is occupied by amino acids in random coils (56.45%-63.69%), followed by β-sheets (20.04%-28.93%) and the lowest proportion is observed in α-helices (9.85%-20.42%). The results of 3D protein structure prediction show that all members of the HmLac gene family possess similar protein structures, suggesting that these proteins may also have similar functions (Figure 3).
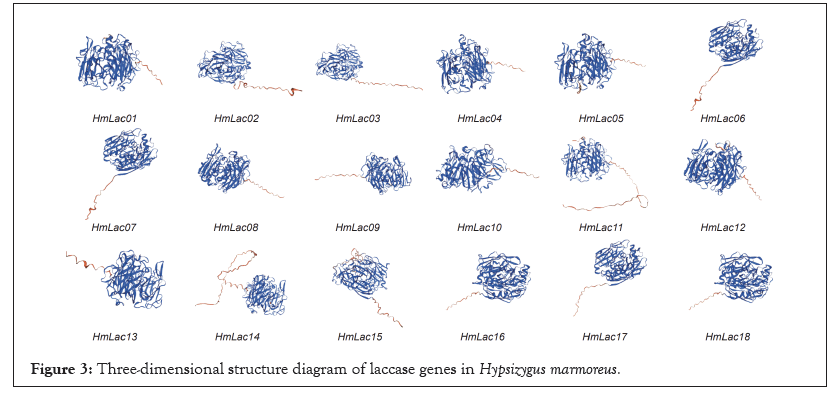
Figure 3: Three-dimensional structure diagram of laccase genes in Hypsizygus marmoreus.
Gene structure, conserved motifs and evolutionary correlations with HmLacs
The comparison of the homology revealed that the consistency of amino acid sequence among laccase genes had a high diversity and the identity ranged from 41.5-90.54%, which might be related to the specificity of functional differentiation in the evolutionary process. Nonmetric Multidimensional Scale (NMDS) analysis of the phylogenetic branch length showed that the white strain was divided into three groups, with significant differences among the groups. The construction of the phylogenetic tree for laccase genes in the HM62-W strain revealed that the genes were classified into four subfamilies. Group 1 comprised of 9 family members, Group 2 consisted of 5 family members, Group 3 included 3 family members and Group 4 contained 2 family members (Figure 4).
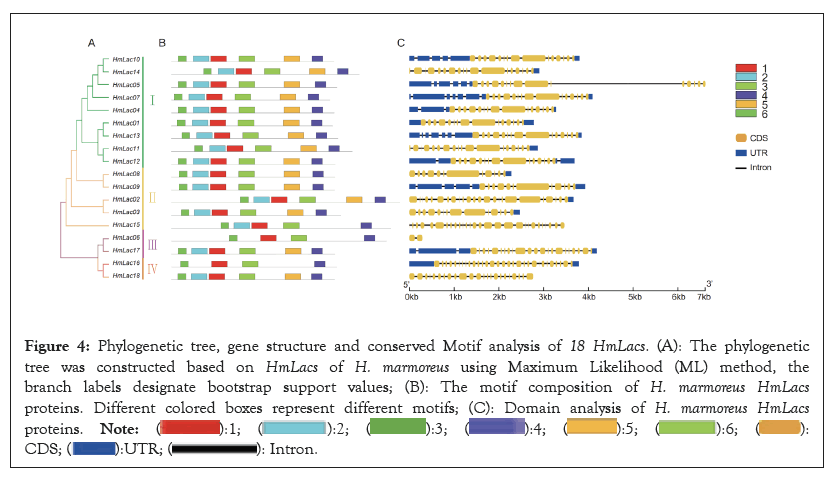
Figure 4: Phylogenetic tree, gene structure and conserved Motif analysis of 18 HmLacs. (A): The phylogenetic
tree was constructed based on HmLacs of H. marmoreus using Maximum Likelihood (ML) method, the
branch labels designate bootstrap support values; (B): The motif composition of H. marmoreus HmLacs proteins. Different colored boxes represent different motifs; (C): Domain analysis of H. marmoreus HmLacs proteins. 
Gene structure analysis showed that the number of exons of the laccase gene ranged from 9-30 and that the structures in each subfamily were similar. In Group 1, 9 Laccase genes had 15 exons. In Group 2, all members except HmLac15 contain 15 exons, suggesting that HmLac15 may have undergone new functional differentiation. HmLac06 in Group 3 is the most unique, with only 2 exons. The laccase genes in Group 4 have the highest number of exons, with 23 and 30 exons, respectively. The number of introns in Group 4 increased significantly, especially in the copper ion binding region (Cu-oxidase_1, Cu-oxidase_2, Cu-oxidase_3), suggesting that the introns might underlie the gene structure diversity and the members of each group may perform different functions. Further analysis showed that all HmLacs genes contain at least one intron in their respective copper ion domain, which may be the conserved intron of the laccase gene. The Motif-based sequence analysis tool was used to identify 6 conservative motifs (length 29-50 aa) of proteins. The comparison of 6 conserved motifs in the CDD database revealed that these motifs were conserved across genes, providing favorable evidence for grouping HmLacs. Among the 18 laccase genes, all except for HmLac15, HmLac06 and HmLac16 contain 5 motifs, while HmLac15 contains 4 motifs and HmLac06 and HmLac16 contain 3 motifs. This observation indicates a strong conservation of laccase genes.
Phylogenetic analysis and classification of HmLacs
To further categorize and investigate the evolutionary correlation of HmLacs, we identified 122 laccase domains from 64 H. marmoreus genomes which assembled by de novo using Whole Genome Sequencing (WGS) data. Based on the classification of HmLacs and the primary structural features of HmLac proteins, all 122 HmLacs genes were classified into four major groups and further divided into seven subgroups; the structure of each group of genes was similar (Figure 5).
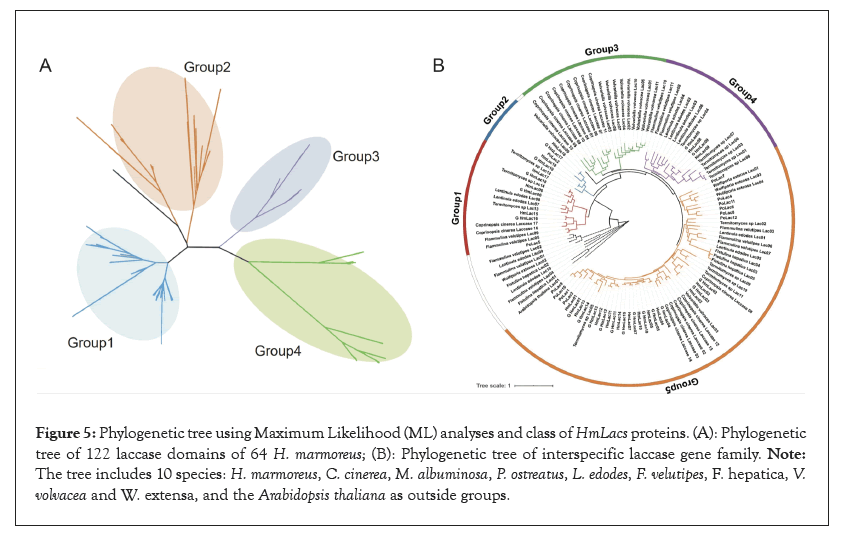
Figure 5: Phylogenetic tree using Maximum Likelihood (ML) analyses and class of HmLacs proteins. (A): Phylogenetic tree of 122 laccase domains of 64 H. marmoreus; (B): Phylogenetic tree of interspecific laccase gene family. Note: The tree includes 10 species: H. marmoreus, C. cinerea, M. albuminosa, P. ostreatus, L. edodes, F. velutipes, F. hepatica, V. volvacea and W. extensa, and the Arabidopsis thaliana as outside groups.
The representative species of basidiomycetes, ligneous white-rot fungus (C. cinerea, M. albuminosa, P. ostreatus, L. edodes and F. velutipes), ligneous brown rot fungus (F. hepatica and W. extensa) and rotting straw fungus (V. volvacea) were used to construct the phylogenetic trees, the Arabidopsis thaliana as outside groups. The results showed that the laccase genes are mainly divided into five groups and each species laccase genes are clustered together respectively, indicating that there were gene replication events. Previous studies suggested that PoLac2 was involved in lignin degradation and fruity body formation. In Group 2, PoLac2 was clustered with HmLac16 and HmLac18, suggesting that the gene might be involved in lignin degradation. Group 3 did not consist of a laccase gene of H. marmoreus. The HmLacs genes are similar to the laccase genes in M. albuminosa and P. ostreatus, but unrelated to those of F. hepatica, W. extensa and V. volvacea.
Expression profiles of HmLac genes in H. marmoreus
The life history of H. marmoreus can be divided into mycelium, mycelium kink (discoloring), primordial, bud and forming stages. In the present study, the samples of the grey and white mycelium of H. marmoreus in the post-ripening, kink (5 days after cap opening) and primordium formation stages (8 days after cap opening) were subjected to transcriptomic sequencing analysis.
Previous studies speculated that the white strain was the albino strain of the gray strain, which was weaker than the gray strain in growth speed and stress resistance and the expression levels of the three transcriptomes of H. marmoreus was recorded (Figure 6). In the transcriptomic experiment, HmLac09 and HmLac10 were highly expressed in the gray strain, white mononuclear mycelia and binuclear heterozygous mycelia, followed by HmLac14, suggesting that these genes are related to the growth and development of H. marmoreus. The expression levels of HmLac02, HmLac05, HmLac08 and HmLac17 genes in gray or heterozygous strains were higher than those in the white mononuclear strains but did not differ significantly compared to the other laccase genes. In the cap epidermal transcriptome experiment, HmLac09 gene was highly expressed in all three strains. The expression of HmLac02, HmLac03, HmLac17, HmLac14 and HmLac18 in gray and hybrid was higher than that of the white strain, while the expression of HmLac10 in white and hybrid was higher than that of the gray strain. The other 11 laccase genes were low expression, suggesting that they played a small role in the development of the cap. The results of transcriptome experiment III showed that HmLac09 and HmLac10 were highly expressed in different periods, further proving the critical role of these two genes in the growth and development of H. marmoreus. The expression levels of HmLac02, HmLac05, HmLac08 and HmLac17 genes in gray or heterozygous strains were higher than those in white mononuclear strains and formed differential expression, which was consistent with the results of experiments I and II, indicating that any one of HmLac02, HmLac05, HmLac08 and HmLac17 may be a factor leading to different growth rates of the gray strain in lignin decomposition. Taken together HmLac09, HmLac10, HmLac17 and HmLac18 are the key laccase genes in H. marmoreus.
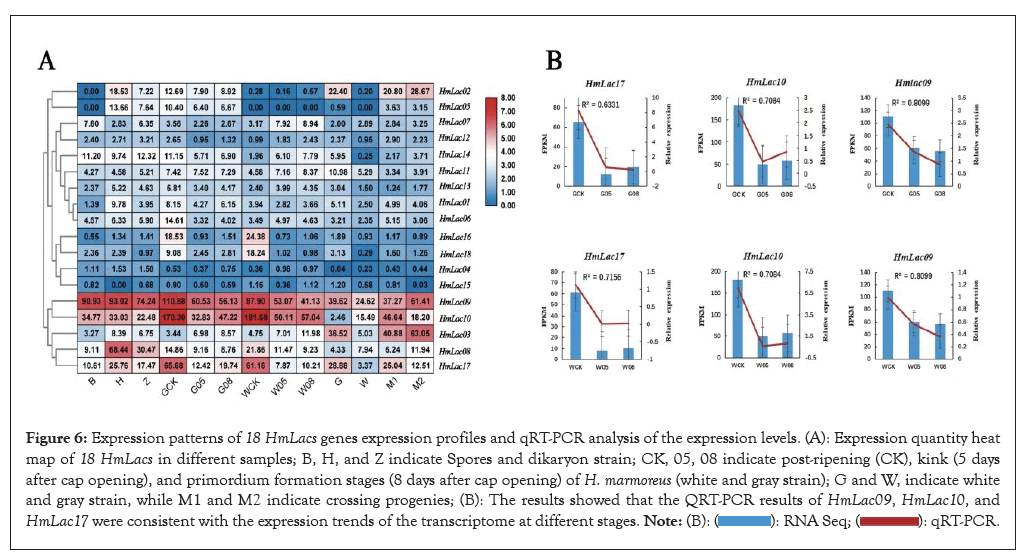
Figure 6: Expression patterns of 18 HmLacs genes expression profiles and qRT-PCR analysis of the expression levels. (A): Expression quantity heat map of 18 HmLacs in different samples; B, H, and Z indicate Spores and dikaryon strain; CK, 05, 08 indicate post-ripening (CK), kink (5 days after cap opening), and primordium formation stages (8 days after cap opening) of H. marmoreus (white and gray strain); G and W, indicate white and gray strain, while M1 and M2 indicate crossing progenies; (B): The results showed that the QRT-PCR results of HmLac09, HmLac10, and HmLac17 were consistent with the expression trends of the transcriptome at different stages. 
The expression of HmLac17, HmLac10 and HmLac09 in different color strains was verified by Real-Time quantitative Reverse Transcription (qRT-PCR). These results showed that the correlation coefficients between Fragments Per Kilobase of exon model per Million mapped fragments (FPKM) and QRT-PCR were R²=0.7312(0.6331), R2=0.7084 (0.7084) and R2=0.8099 (0.8099) for HmLac17, HmLac10 and HmLac09, respectively. The results showed that the QRT-PCR results of HmLac09, HmLac10 and HmLac17 were consistent with the expression trends of the transcriptome at different stages. The expression levels of these three laccase genes were higher in the post-ripening of the gray and white strains than in the kink (5 days after cap opening) and primordium formation stages (8 days after cap opening).
Laccase of white-rot fungi is a major lignin-degrading gene that has been widely studied in recent years due to its high lignin-degrading efficiency. As one of the typical white-rot fungi, its nutritional components mainly originate from the degradation of lignin, hence, the laccase activity directly affects the production efficiency of H. marmoreus in the factory. Based on whole genome identification, several laccase genes have been identified in many edible mushroom species, such as P. ostreatus and F. velutipes. In this study, the high-quality genome of H. marmoreus was utilized to compare the homologous sequences of species previously published at the whole genome level. Additionally, the SMART database, CDD database and laccase marker (L1-L4) were employed to identify 18 laccase family genes. This finding, which identified 18 laccase family genes, was 8 more than the 10 laccase genes previously identified by Zhang, et al through transcriptome assembly and splicing [16]. These 8 genes contain the characteristic structure of the copper ion binding region and are accurate members of the laccase gene family, which consists of 18 members. Currently, it is the largest laccase gene family found in Basidiomycota, exceeding the 17 laccase genes in C. cinerea, which might be the result of the growth and development of H. marmoreus and its dependence on laccase.
Based on the IEPs of the laccase gene family members, it was identified that HmLac2 is a basic protein. H. marmoreus has a tendency to grow in an acidic environment, yet basic substances are secreted during its growth process, gradually increasing the pH value of the culture material. HmLac2 plays a crucial role during this specific period. Additionally, the mushroom possesses both acidic and basic proteins, indicating differences in its acid-base stability. This finding is consistent with the previous conclusion that fungal laccase exhibits a wide optimal pH range [36]. Similar situations have also been observed in other species, for example, PoLac8 in P. ostreatus, which necessitates further experiments to verify their specific functions. Some studies proposed that laccase was a secreted protein, but the results of signaling peptides in this experiment showed that some laccase did not have typical signal peptides, as in F. velutipes, V. volvacea and L. edodes. Also, laccases might function as intracellular enzymes with specific roles. Thus, we hypothesized that laccases, such as HmLac2, HmLac6, HmLac7, HmLac11, HmLac14 and HmLac15 may act as extracellular proteins without typical signaling peptides.
The distribution of 18 HmLacs on chromosome 6 was not uniform and 9 genes were clustered linearly on chromosome 6, indicating that the laccase gene was derived from gene replication. The main driving force of the laccase gene family expansion was tandem repetition and reverse transcription transpose; 7 tandem repetition genes were identified indicated that the original laccase genes were differentiated into paraphyletic homologs with different functions during the evolution process to meet the various functional requirements of fungi throughout the life cycle. Six lineal homologs have been identified in H. marmoreus and P. ostreatus and it has been inferred that these genes may come from a common ancestor. The results of gene structure analysis showed that most HmLacs genes enriched in the same group had similar intron numbers. The number and distribution of introns were related to gene evolution, which could be attributed to intron insertion or deletion caused by environmental pressure after species differentiation. In order to respond to various stresses promptly, genes must be activated quickly. In this respect, compact gene structures with fewer introns are conducive to expression [37].
Phylogenetic analysis of laccase genes among different species indicates that the 122 HmLacs were divided into five groups. Within each group, the laccase genes clustered together, suggesting that the formation of laccase genes predates speciation and that gene replication events occurred after species differentiation. The laccase gene family of H. marmoreus is closely related to L. edodes and F. velutipes, while most V. volvacea and C. cinerea genes cluster in a single branch. This indicates that the laccases of H. marmoreus are closely related to wood rot fungi but more distantly related to grass rot fungi, aligning with previous findings [38].
Laccases play a crucial role in fungi as they are involved in various physiological and metabolic processes and involved in lignin degradation and the synthesis of fungal pigments, such as melanin. For example, PoLac2 was overexpressed in P. ostreatus by Agrobacterium-mediated transformation, the expression level of the PoLac2 gene in the transformant was 2-8 times higher than that of the wild type and the lignin degradation rate of the transformant was 2.36-6.3% higher than that of the wild type within 30 days. In this study HmLac09, HmLac10 and are highly expressed at various developmental stages and in the mononuclear mycelium and hybrid seed. Particularly, the expression levels of HmLac16, HmLac17 and HmLac18 reached their peak in the early bag-opening stage (post-ripening stage), which is a period of vigorous nutritional accumulation. This indicates that these three genes play a significant role in the degradation of lignin and provide nutrients for growth. Moreover, they are crucial genes in the laccase gene family involved in lignin degradation. Moreover, HmLac17 were clustered on the same branch as PoLac2 in the Phylogenetic tree of laccase gene, as previously reported, PoLac2 plays an important role in the decomposition of laccase.
Conceptualization, H-BL, B-BL, FL and GW; methodology, XX-W and YC; formal analysis, Y-YW and H-BW; investigation, B-BL; data curation, H-BW and GW; writing-original draft preparation, GW, B-PT and LM; writing-review and editing, YC, CW and GW. All authors have read and agreed to the published version of the manuscript.
This study was supported by the Open Foundation of Jiangsu Key Laboratory for Bioresources of Saline Soils [JKLBZ202005], Jiangsu Province industry-university-research cooperation project [BY2021457] and National Natural Science Foundation of China [32002108].
The original genome data was uploaded to NCBI BioProject, under the accession number: PRJNA508399.
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript or in the decision to publish the results.
The genome sequences of H. marmoreus have been deposited at GeneBank under the accession number of JABWDO000000000. The data from this study were deposited with NCBI GenBank under accession numbers: PRJNA508399 and PRJNA644211
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Wang G, Liu R, Wang H, Sun X, Wu X, Zhu Y, et al (2023) Genome-Wide Identification and Characterization of the Laccase Family in Hypsizygus marmoreus and Expression Profile Analysis. Fung Genom Biol. 13:233.
Received: 21-Nov-2023, Manuscript No. FGB-23-28099; Editor assigned: 23-Nov-2023, Pre QC No. FGB-23-28099 (PQ); Reviewed: 08-Dec-2023, QC No. FGB-23-28099; Revised: 15-Dec-2023, Manuscript No. FGB-23-28099 (R); Published: 22-Dec-2023 , DOI: 10.35248/2165-8056.23.13.233
Copyright: © 2023 Wang G, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Sources of funding : This study was supported by the Open Foundation of Jiangsu Key Laboratory for Bioresources of Saline Soils [JKLBZ202005], Jiangsu Province industry-university-research cooperation project [BY2021457] and National Natural Science Foundation of China [32002108].