Drug Designing: Open Access
Open Access
ISSN: 2169-0138
ISSN: 2169-0138
Research Article - (2020)Volume 9, Issue 2
Starting from the concept of "similar molecules exert similar biological activities" to modify structures of biologically active molecules or readily available drugs to improve their therapeutic activity or reduce side effects. Four designed 7- Chloroqunioline-N-Heterocyclic molecules that are derived from parent compound chloroquine (which recently has attracted wide international attention due to its potential activity against COVID-19) were investigated regarding QSAR and some electronic parameters compared to the original drug. Their chemical properties, biochemical properties, and physio-chemical properties were evaluated using computational chemistry. The method used for calculations was semi-empirical-geometry optimization-PM3. Single crystal X-ray crystallography data of Chloroquine and calculated data have been done using the same parameters. Surprisingly, the experimental and calculated data recorded the same values. Furthermore, in Silico evaluation of pharmacokinetics, drug-likeness and medicinal chemistry friendliness of four hybridized molecules compared to chloroquine by using SwissADME web tool confirm closeness of four hybrids to the chloroquine. The total energy, binding energy, dipole moment energy gap (HOMOLUMO) and log p of one of compounds coded AB1-4Py were -7.6647.710 Kcalmol-1, -3981.323, 4.531, 8.161 eV, and 4.25 respectively. AB2-4py compound calculated total energy was 76007.6- , Binding energy at - 4109.424, dipole moment at 4.464, energy gap (EHOMO-ELUMO) at 8.131 and log p at 4.70. The AB1-4py, AB2-4py and chloroquine reported same stability and bioactivities. From QSAR study, the value of Log P indicates hydrophobic nature. The recorded values of AB1-4Py and AB2-4py were completely agreed with those of chloroquine. Therefore, this study may valuable in the discovery of a new series of potential drugs for treatment of malaria or COVID-19.
Chloroquine; COVID-19; Computational; Anti-viral; Hybridized Molecules; Medicinal chemistry
In December 2019, the outbreak of novel coronavirus that also known as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2 or COVID-2019) was first reported in Wuhan, China. Since that, it widely spread in China as well as many other countries. Therefore, many research laboratories, clinical trials centers and cell culture studies are still ongoing trying to control the rapidly spreading and life threatening viral infection that can cause pneumonia induced death in approximately 2.5% of infected individual according to the latest report in March 14th 2020.At present, researches have proven that chloroquine may inhibit the growth of SARS-CoV-2 20. This finding has been supported by clinical studies conducted in approximately one hundred SARS-CoV-2 infected patients [1-5].
Chloroquine a 70 years old drug that has been used as prophylactic and therapeutic anti-malarial drug in endemic areas. Additionally, as daily treatment for autoimmune diseases, attracted wide attention due to the therapeutic efficacy it might have against COVID-19. The drug has been included officially in federal guidelines for treatment of COVID-19 in Republic of China with many cautions due to its serious side effects [6,7].
Many studies suggest that chloroquine may exert its antiviral activity through inhibition of pH-dependent viral fusion/ replication and prevention of viral envelope glycoprotein as well as host receptor protein glycosylation. The drug may also inhibit virion assembly in Endoplasmic Reticulum-Golgi Intermediate Compartment (ERGIC)-like structure. In host the drug can exert its action by attenuation of the expression of pro-inflammatory factors that cause acute respiratory distress syndrome; the first cause of most of deaths in COVID-19 patients [8].
Actually; in new ages of medicinal chemistry and drug design the computational chemistry; molecular modeling and automated synthesis are utilized extensively by researches to help design diverse libraries of new drugs with more potency and less side effects. So, many antiviral drugs like Ritonavir, Indinavir, Zanamivir, Oseltamivir, Lopinavir and others have been discovered by computer-based methods. Therefore, these tools are considerable to be the working horse in many pharmaceutical companies help to resolve many global crises.
CLOGP is one of the most important program that was launched in 1980s and could predict the lipophilicity that could be related to predication of some molecular properties such as molecular volume, molecular surface area, transport through membranes, and binding to receptor surfaces, and hence to many different bioactivities. However, bioactivity of molecules can be estimated using computational Quantitative structureactivity relationship (QSAR) as well as some of their electronic structure such as HOMO/LUMO energy gap [9].
QSAR have been used to model most of the common biological activity of molecules such as bio concentration factor,biodegradation rate, carcinogenicity,drug absorption, drug metabolism, drug clearance,inhibition constant, mutagenicity, blood brain barrier (BBB) permeability, skin permeability,pharmacokinetics,receptor binding affinity,solubility and toxicity [10-23].
In addition, hybridized molecules especially with N-heterocyclic moieties are considered to be one of the most important classes of nitrogen containing heterocyclic compounds that exhibit various biological activities and have many applications in medicinal chemistry. Qunionline-triazoles molecules are good examples of these hybridized molecules that offer treatments for many types of diseases such as cancer and others [24-26].
In the present research, electronic properties and the QSAR studies were performed for four N-heterocyclic hybridized molecules in order to investigate the quantitative effect of the various Physico-Chemical parameters of four compounds closed to chloroquine. Two of the set hybridized molecules recorded values identical to that of chloroquine. In Silico evaluation of pharmacokinetics, drug-likeness and medicinal chemistry friendliness of four hybridized molecules compared to chloroquine by using SwissADME web tool confirm closeness of four hybrids to the chloroquine. Suggesting that a new series of drug could be synthesized to replace chloroquine [27].
Personal acer desktop PC with Windows XP, a 2.93 GHz Intel (R) core (TM) 2 Duo CPU, and 4.00 GB of RAM was used to run all the calculations. Semi-empirical PM3 quantum chemical calculation was carried out by the HyperChem (version 8 ) TM 8.0 Molecular Modeling program with root mean square (RMS) gradient 0.1 kcal/Ǻmol using PolakRibiere algorithm. The calculations for all compounds runs using semi-empiricalgeometry optimization- PM3 methods. Physiochemical prediction of hybrid compound was performed by using SwissADME web tool [28-31].
Considerable attention should be given to the substituted quinoline ring specially in chloroquine as suggested treatment of COVID-19 pendamic. Quinoline ring is class of N-Heterocyclic compounds whose used widely as base for many of medications such as Moxifloxacin, Grepafloxacin, Ciprofloxacin and Cinchocaine. The presence of high electronegativity atom such as chloride in quinoline ring will affect bioactivity of the compound. Moreover, the N-heterocyclic rings such as qunioline environmental sensitive. So, the substitutions in quinoline ring may increase the bioactivityof molecules as well as increase their intermolecular interactions. The π-π-stacking between quinoline rings in anti-malarial treatments have been reported as one of the most important non-covalent interactions.Additionally, this kind of interactions reported as one of the most important interactions that increased molecular stability. As well as plays an important role in intermolecular recognition [32-40].
However, this interactions may include intercalation of DNA. Moreover, relevant noncovalent interaction patterns responsible for intermolecular recognition may enhance the biological functionality of our designed compounds.Recognition or selfassembly can be described as ‘a process in which two or more components can spontaneously associate to form a larger, moreordered structure through non-covalent interactions [41].
However, the hybridized molecules may increase their biofunctionality [26] through their components using weak interactions reversible assemble. Self-assembly here is generally driven by relatively weak interactions which are often electrostatic and can include π-π-stacking [26,41,42].
Tobacco mosaic virus (TMV) and protein sub-units spontaneously arrange around single strand of RNA to form a right handed helix with 16 ⅓ sub-units per helical turn were the natural examples of strict self-assembly [43].
Semi-Empirical-Geometry Optimization-PM3
The hybridized molecules offer better treatment for different types of diseases.Molecules building their framework based on quinolone-N-Heterocyclic moieties (see Figure 1); can be classified as hybridized compounds. Despite the qunioline as base tend to be one of the most important unit in our life molecules due to its functionality in the life organism; its combination with bioactive moieties may produce a new products with new functionality. Some QSAR parameters and some electronic properties of the hybridized molecules will be discussed below [25,26,44].
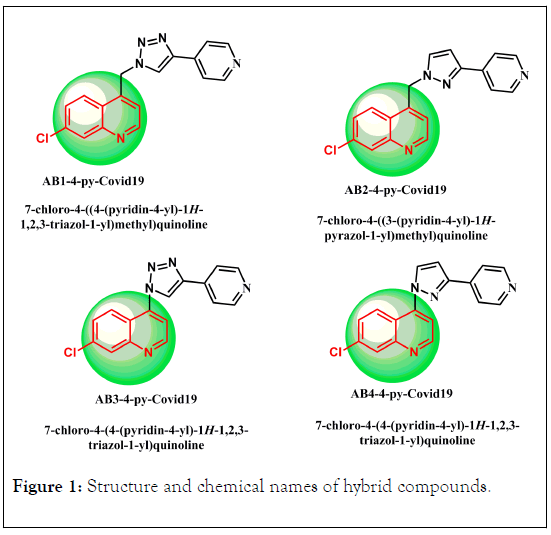
Figure 1: Structure and chemical names of hybrid compounds.
3D view of molecule AB1-4py and AB2-4py were presented in Figures 2 and 3 respectively. The HOMO, LUMO orbitals is very important to mention the chemical reactivity and region of chemical reactivity as well as indicate where the electrophilic and neucleophilic can be attached. The calculation values for all molecules were reordered in Table 1. The observed results that closed to the chloroquine recorded for compounds coded AB1-4py and AB2-4py. AB1-4py compound recorded total energy at -7.6647.710, binding energy at -3981.323, dipole moment at 4.531, energy gap (HOMO-LUMO) at 8.161 and log p at 4.25. Whereas, AB2-4py registered total energy, binding energy, dipole moment energy gap (HOMO-LUMO) and log p at -76007.626, -4109.424, 4.464,8.131 and log p 4.7 respectively. The AB1-4py, AB2-4py and chloroquine recorded same energy gap values. As the molecules stability can be measured based on the EHOMO/LUMO gap and all of AB1-4py, AB2-4py and chloroquine present same values; which main that the molecules have same stability. Moreover, the bioactivity of molecules can be estimated by log p;AB1-4py, AB2-4py and chloroquine recorded same values of lop p which is at 4; that reflect the three molecules may have same bioactivity [45-47].
| Parameter | AB1-4Py | AB2-4Py | AB3-4Py | AB4-4Py | Chloroquine |
|---|---|---|---|---|---|
| Total energy Kcal/mol | -7.6647.710 | 76007.626- | 73196.227- | 72558.344- | -76971 |
| Binding energy Kcal/mol | -3981.3 | 4109.424- | 3697.868- | 3828.1703- | -4788.2 |
| Dipole moment (deby), D | 4.531 | 4.464 | 3.063 | 3.25 | 4.097 |
| EHOMO(eV) | -9.505 | -9.301 | -9.329 | -9.069 | -8.9217 |
| ELOMO(eV) | -1.344 | -1.178 | -1.624 | -1.438 | -0.7871 |
| Energy gap(eV) | 8.161 | 8.131 | 7.705 | 7.631 | 8.1345 |
| Surface area (Approx) | 398.63 | 413.83 | 374.49 | 384.15 | 568.85 |
| Surface Area (Grid) | 532.54 | 543.54 | 497.18 | 500.53 | 596.39 |
| Volume | 882.72 | 895.65 | 826.33 | 834.03 | 1007.6 |
| Polarizability | 35.95 | 36.66 | 34.12 | 34.82 | 38.14 |
| Reactivity | 90.06 | 90.2 | 85.23 | 85.23 | 96.39 |
| Log P | 4.25 | 4.3 | 4.15 | 4.6 | 4.27 |
Table 1: Data of ABn-4Py (n=1,2,3,4) Semi-empirical-PM3-geometry optimization Conclusion.
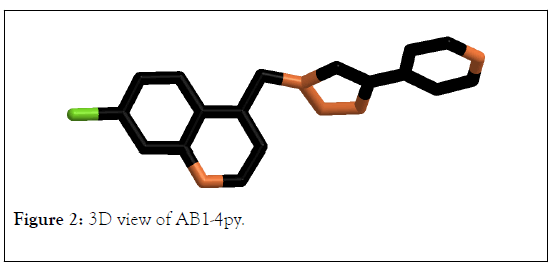
Figure 2: 3D view of AB1-4py.
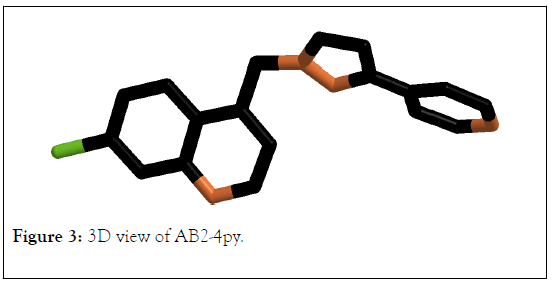
Figure 3: 3D view of AB2-4py.
Geometrical investigation of Chloroquine as reference compound
The crystal structure of chloroquine with atomic number presented in Figure 4. Bond length (Å) for experimental and calculated data. The recorded values of both crystal and calculated were closed as we can see that in Table 2. However, selected bond angle (˚) presented in Table 3. The experimental and calculated values seem in excellent agreement. The single crystal values of bond angles and bond lengths in good agreement with calculated values [38].
| Experimental Bond length | Calculated Bond length | Experimental Bond angles | Calculated Bond angles | ||
|---|---|---|---|---|---|
| N(I)-C(2) | 1.323 (6) | 1.32303 | C(15)-C(16) | 1.499 (7) | 1.51784 |
| C(4)-N(9) | 1.334 (6) | 1.44267 | C(l0)-C(19) | 1.513 (7) | 1.52561 |
| N(9)-C(10) | 1.481 (6) | 1.49727 | C(3)-C(4) | 1.403 (6) | 1.3787 |
| C(13)-N(14) | 1.485 (6) | 1.49085 | C(17)-C(18) | 1.498 (8) | 1.51639 |
| N(14)-C(17) | 1.495 (6) | 1.4919 | C(2)-C(3) | 1.360 (6) | 1.41881 |
| N(14)-C(15) | 1.511 (6) | 1.49517 | C(1 l)-C(12) | 1.516 (6) | 1.52171 |
| C(12)-C(13) | 1.513 (6) | 1.52424 | CI-C(7) | 1.723 (5) | 1.68559 |
Table 2: Selected experimental and calculated bond lengths (Å).
| Calculated Bond angles | Experimental Bond angles [38] | Calculated Bond angle | Experimental Bond angle [38] | ||
|---|---|---|---|---|---|
| C(4)-C(3)-C(2) | 120.6 (5) | 119.75 | C(8)-C(7)-Cl | 120.1 (4) | 120.149 |
| N(9)-C(4)-C(3) | 121.4 (5) | 120.684 | C(6)-C(7)-Cl | 118.7 (4) | 118.479 |
| C(10)-N(9)-C(4) | 124.7 (4) | 115.555 | C(15)-N(14)-C(13) | 108.3 (4) | 113.747 |
| C(12)-C(11)-C(10) | 113.9 (4) | 110.634 | C(18)-C(17)-N(14) | 115.6 (5) | 111.27 |
| N(14)-C(13)-C(12) | 114.3 (4) | 110.857 | C(3)-C(2)-N(1) | 122.4 (5) | 122.373 |
| C(17)-N(14)-C(13) | 114.7 (4) | 112.099 | C(19)-C(10)-C(11) | 112.0 (4) | 110.439 |
| C(16)-C(15)-N(14) | 114.6 (4) | 110.139 | C(I I)-C(10)-N(9) | 109.4 (4) | 108.388 |
Table 3: Selected experimental and calculated bond angle (˚).
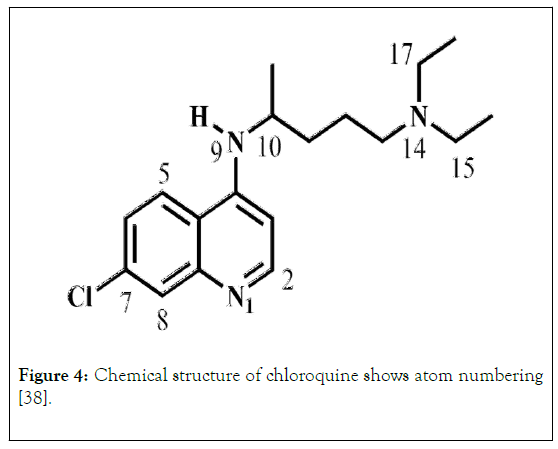
Figure 4: Chemical structure of chloroquine shows atom numbering [38].
Physicochemical, pharmacokinetic, medicinal chemistry and drug likeness prediction
To qualify the proposed structures as candidates, the compounds molecular properties of Lipinski's rule of five were calculated using SwissADME web tool. The number of hydrogen bond acceptors and donors for all compounds were in accordance with Lipinski`s rule of five. The molecular weights of all compounds were within the range [48,49]. The lipophilicity property [50] (MLogP ≤ 4.15, octanol-water partition coefficient) was in the range for all compounds and results are shown in Table 4.
| Compounds | Mwta | MLogPb | nHBAc | nHBDd | n violation sf |
|---|---|---|---|---|---|
| Rule | <500 | < 4.15 | <10 | <5 | 0 |
| AB1--Py | 321.76 | 2.23 | 4 | 0 | 0 |
| AB2-44-Py | 320.78 | 2.48 | 3 | 0 | 0 |
| AB3-4-Py | 307.74 | 2.26 | 4 | 0 | 0 |
| AB4-4-Pv | 306.75 | 2.51 | 3 | 0 | 0 |
| AB4-4-Py | 306.75 | 2.51 | 3 | 0 | 0 |
aMolecular Weight; bCalculatedLipophilicity (MLog Po/w); cNumber of Hydrogen Bond Acceptors.dNumber of Hydrogen Bond Donors; eViolations from Lipinski’s Rule.
Table 4: Lipinski`s rule of five for the four hybrids compounds.
SwissADME also has computational filters that include Ghose, Egan,Veber, and Muegee developed by leading pharmaceutical companies and cheminfomaticians to evaluate the drug-likeness based on several parameters; Ghose evaluation by computing physicochemical property, presence of functional groups and important substructures, Egan predict the drug absorption based on physical processes involved in membrane permeability, Veber (GSK filter) model characterizes molecules as drug-like if they have 10 or fewer rotatable bonds and a PSA (polar surface area), Muegge (Bayer filter) model is a database-independent pharmacophore point filter that discriminates between drug-like and nondrug-like chemical matter. ketone, hydroxyl, sulfonyl and amine groups are considered to be the most important four functional motifs in drug-like molecules included in testing for all above mentioned filters;the four hybrids shows close likeness to the chloroquine with good scores the data are shown in supplementary I [49].
Finally, the medicinal chemistry was evaluated and result shown in Table 5 confirmed the likeness of the four hybridized molecules to chloroquine with acceptable synthetic accessibility.
| Compounds | PAINS Alert | Brenk Alert | Lead Likeness | Synthetic Accessibility |
|---|---|---|---|---|
| AB1-4-Py | 0 | 0 | Yes | 2.57 |
| AB2-4-Py | 0 | 0 | Yes | 2.36 |
| AB3-4-Py | 0 | 0 | Yes | 2.65 |
| AB4-4-Py | 0 | 0 | Yes | 2.32 |
Table 5: Medicinal chemistry evaluation of hybrids.
Finally, Swiss ADME program evaluated the Pharmacokinetic properties of hybrids like, gastro-intestinal absorption, blood brain barrier permeability, inhibition the cytochrome P450 isoenzymes (CYP isoenzymes), and substrate P-glycoprotein gas demonstrated below in Table 6 all hybrids presented (yes) to CYP and all could substrate Pgp except compound coded AB3-4- Py [48].
| Compounds | GI absorption | BBB perment | Pgp Substrate | CYPX Inhibition |
|---|---|---|---|---|
| AB1-4-Py | High | Yes | Yes | Yes |
| AB2-4-Py | High | Yes | Yes | Yes |
| AB3-4-Py | High | Yes | Yes | Yes |
| AB4-4-Py | High | Yes | Yes | Yes |
Table 6: Pharmacokinetic evaluation of hybrids (GI: Gastro-Intestinal Absorption, BBB: Blood Brain Barrier, CYP: Cytochromes, P-gp: PGlycoprotein.
Four molecules and chloroquine investigated using semiempirical, geometry optimization and PM3 calculation methods and In-silico evaluation. Geometrically, the single crystal X-ray crystallography of chloroquine revealed that the data of bond lengths and bond angles completely agreed with calculated.
The two compounds AB1-4Py and AB2-4Py recorded bioactivity and electronic properties that are very close to the chloroquine. The N-heterocyclic units of molecules seem to be more flexible which may enhance the functionality of products. The two molecules and chloroquine; recorded same values of energy that is why the entire compound has same stability [49-51].
The bioactivity of molecules can be estimated by log p. AB1-4py, AB2-4py and chloroquine recorded same values of lop p which is 4; that reflect the three molecules have same bioactivity. Furthermore, we used the SwissADME virtual tools to further evaluate the medicinal chemistry, drug likeness, pharmacokinetics and physiochemical properties the results have proven closeness to chloroquine, hence these molecular hybrids are especially interesting for researchers to working with compounds as same as the chloroquine bioactivity; as antiviral therapies or antimalarial and may for COVID-19. Ultimately, biological evaluation and in vivo studies are required to validate the pharmacokinetics and pharmacodynamics of any potential patient drug.
We thank Ministry of Education, University of Benghazi and Mr. Abdulrazeg H. Elnathoury the head of COVID-19 Scientific Committee for their assistance.
Citation: Najar AM, Omar RMK, Bobtaina E (2020) Computational, Pharmacological Evaluation and Comparative Similarity Against Chloroquine for Some New Designed Hybridized Molecules and Their Potential Use as Antiviral Against COVID-19 and Malaria. Drug Des. 9:163.
Received: 24-May-2020 Accepted: 08-Jun-2020 Published: 15-Jun-2020 , DOI: 10.35248/2169-0138.20.9.163
Copyright: © 2020 Najar AM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.