Journal of Antivirals & Antiretrovirals
Open Access
ISSN: 1948-5964
ISSN: 1948-5964
Research Article - (2019)Volume 11, Issue 2
Recently 2018, four strains of infectious bronchitis virus (IBV) were isolated from vaccinated and non-vaccinated broiler flocks from different chicken farms in different cities in Iraq. The pathogenic characters were assessed for the four IBV strains. Test chickens were infected by the four strains releasing an immune response. Noticeable results showed by cross neutralization assays which revealed that strains antigenically distinct from classic IBV strains of H120, M41, Conn, and Gray. Compared to H120 vaccine strain, short insertion, point mutation, and deletion occurred at many positions in the S1 protein of the four strains. Two of the four strains had the motif (HRRRR), which was considered identical to that of the epidemic IBV strains in Iraq. Another two strains had new motifs (HRLRR and RRYRR) emerged in the isolated strains. These Four strains were also genetically different from the vaccine strains and non-Iraqi IBV strains but closely related to large numbers of USA and China, Singapore and India strains. The isolates and 14 reference IBV strains were clustered into three distinct groups (I-III). The four strains were categorized into groups III, forming a big phylogenetic branch, which is closely related to USA IBVs and Singapore IBVs whereas the vaccine strains belonging to group I and II are genetically distant from groups III. The results from this study indicate that different IBV strains circulate in the chicken population in Iraq.
Infectious bronchitis virus; S1 gene; Phylogenetic analysis
Corona virus is the main cause of avian infectious bronchitis (IBV) [1] which is an acute and highly contagious disease of chickens. The main clinical sings that could appear through the infection is tracheal rales, respiratory system, including gasping, coughing, sneezing and nasal discharge. Respiratory distress with decreasing in the egg production has been reported due to the infection in laying fowls [2]. In addition, kidney lesions were reported by some researcher [1,3] IB is one of the major problems in poultry industry in many countries which has caused significant economic loss. For that, serological and molecular characterization of IBVs isolates very important. Coronaviridae the family which IBV belongs to it and genome consists of a single-stranded positive sense RNA encoding four structural proteins.
During viral maturation S glycoprotein will transnationally in to S1 and S2 subunits [4]. Previous studies have been mention that one important point was in believing S1 protein had an important role in providing protective immunity. As well as, they mentioned incidence of new serotype which causing tissue tropism due to the mutation in S1 gene in the N-terminal [5,6]. Different IBV serotypes have been identified throughout the world and keep emerging in in spite of vaccination programme which is related to that IBV exhibits extensive anti-genic variation, and each country has unique strains. We isolated four IBV strains from infected chickens on different farms in Iraq (Table 1).
| Strain | Year of isolation | Tissue | Age (days) | Location | Clinical signs | Accession no | Recognition motifs |
|---|---|---|---|---|---|---|---|
| MK562377 | 2018 | Kidney | 10 | Babylon | Respiartory Nephritis |
MK562377 | HRRRR |
| MK562376 | 2018 | Kidney | 27 | Baghdad | Nephritis | MK562376 | HRRRR |
| MK562375 | 2019 | Trachea | 22 | Wasit | Nephritis Respiratory |
MK562375 | RRYRR |
| MK562374 | 2019 | Kidney | 28 | Mesan | Nephritis | MK562374 | HRLRR |
Table 1: IBV strains isolated from flocks in different areas of Iraq.
Between 2001 and 2019; In this study, we mentioned the antigenic and molecular characteristics of these IBV strains depending on the results from chicken embryo crossneutralization assay, virulence test on 10 day old chickens, and sequence analysis of the S1 glycoprotein gene.
Four IBV strains (MK562374, MK562375, MK562376, MK562377) were isolated during (2018-2019) as mentioned in Table 1 with known source viruses four vaccine strains were obtained from Babylon veterinary hospital (H120, M41, Conn and gray). 10 day old healthy chicken embryo (no infection detected) were injected with the viruses in the allantoic cavities, later after 72 hours allantoic fluid was collected and frozen at -8ºC for the next step (RNA extraction), from the pathogenic signs observed on chicken embryo such as curled and Dwarfed embryo IBV was confirmed as well as to the molecular detection (RT_PCR) for N protein gene [7].
Experimental infection
Ten days old chicken were kept in the safe environment (free infection) at Lab animal, our experimental plan was to divided chickens into to five groups and each group included 10 chickens 1-5 group were inoculated respectively with four IBV strains by intraocular instillation (100 μl) of allantoic fluid 1 × 105 as the infective dose (EID50) of the relevant strain control group inoculated with sterile allantoic fluid. After inoculation chickens were examined daily for observation of IBV signs till 28 days then swabs were collected from tracheal on days 4, 8, 12 and 16 each swab was saved individually in 200 μl of medium for virus isolation (50% glycerol and phosphate buffer solution at -8ºC as well as blood sample collected and saved at -8ºC.
Production of antisera against the isolates: Seventeen free infection chickens of 1month of age were divided into four groups, each group were inoculated with the four strains by instillation intraocular with 1 × 104 EID50. Chickens were slaughtered after 20 days and blood collected from all chickens at euthanatization with a 50 ml centrifuge tube ( Costar, Corning, NY ) [8]. The blood sample were kept at room temperature for 12 hours prior to being centrifuged at 7500 x g for 30 minutes to separate the serum. The antibody titers were determined with an enzyme-linked immunosorbent assay ( ELISA) kit (IDEXX, Westbrook, ME). The test sera were diluted 500 times before being analyzed, and the optical density was measured at 650 nm using an ELISA microplate reader. Each serum sample, including negative and positive controls, was analysed in triplicate.
Cross-neutralization assays for the chicken embryo: To reveal the relationship between the classic IBVs trains (HI20, M41, Conn, and Gray) with the our four strains, this test done on 10 day old non infected embryonated eggs, assay was modified from a virus-neutralizationtest procedure described previously [3,8] Serial 10-fold dilutions of I BV strains were prepared ( 10-106 EID50/0.1 ml), and each dilution was mixed with a standard dilution of serum(1/10 dilution) then kept at room temperature for 1 hr. The serum-virus mixtures in each
Dilution was inoculated into the allantoic cavity of 10-day-free infection chicken embryos in five replicates. Eggs were also inoculated with the corresponding titters of virus alone in parallel after inoculation the noticeable characteristic of IBV lesions was dwarfing, stunting of embryos were examined, and the EID50 was determined by the Reed-Muench method [3,8]. The neutralization index N1 as the ratio of virus-serum EID50 to the corresponding titers of virus alone EID50 was calculated.
S1 gene primers for amplifying S1 gene was designed using primer 3 plus software based on the alignment of known sequence of S1 gene, Primers are 5 ’ CACCCTAGAGGTTTGTCTATGCAT-3 ’ and 5 ’ TCCACCTCTATAAACACCCTTT-3 ’ respectively. Using infected allantoic fluid for RNA extraction by RNA kit was used (intron biotechnology IQeasy plus viral RNA extraction handbook Cat. No.17153-ver1.0 Korea.
One step RT-PCR was performed using GeneAmpR PCR system 9700 [9] quantities mentioned in the Table 2.
| Reagent | Volume in μl |
|---|---|
| Buffer (10x) | 2 μl |
| Macl 2 | 2.5 μl (25 mmol/1) |
| dNTP | 2.5 μl (10 mmol/1) |
| Each primer | 0.75 μl (10 mmmol/1) |
| Sterilized water | 10.2 μl |
| RNAase inhibitor | 20 U/μl |
| RT | 50 U/μl |
| Gold Taq polymerase | 5 U/μl |
Table 2: RT-PCR Reaction mixture.
The PCR product were analysed on 1.5% agarose gel, RT-PCR product was (650 bp), next purified by Gel extraction Kit (MEGAquick-spinTM PCR& Agarose gel DNA Extraction system) all manufactured steps followed in the last step 15 μl elusion buffer were added. Purified PCR product subjected to sequencing using the BigDye* Terminator v1.1 Cycle sequencing Kit (Life Technology) PCR product sequence from both direction same primers were used.
Phylogenetic tree
Phylogentic tree were constructed using our four sequences with 14 known sequence published in GenBank Clustal W alignment and MegaX software Version 10.0 [9,10] tree constructed for the S1 gene using neighbor joining method with bootstrap 1000 replicate with mega.
Four IBV strains were Isolated from Babylon, Baghdad, Dywanea and Karbalaa of Iraq during (2018-2019). Table 1 tissue samples were collected from broilers chicken that showed a typical signs for the IBV infection such as respiratory and nephritis included pale kidneys and enlarged as well as urate deposited in the tubules, loss of fluid due to dehydration with the weight loss.
Trachea, lung and kidney were collected from the four isolate of IBV strain. A noticeable signs were observed like dwarf and dead embryos in different passages when each isolate was inoculated in to embryos. Serological and molecular (conventional RT-PCR technique) were showed all isolates free of other agent such as (Newcastle disease virus and avian influenza virus. All four isolated strains were verified by the observation of curled and embryos and RT-PCR analysis for N gene of IBV.
Experimental infection of chicken
Four days after inoculation typical signs investigated on treated group this manifestation like respiratory signs, ruffled feather with dark shrunken comb, gross lesion mainly found in kidney parenchyma (swollen and pale) with mottled tubules, chessy material in bifurcation of bronchi as shown in Figure 1 as well as urethras were distended with uric acid crystal.
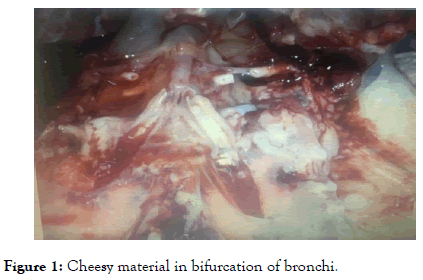
Figure 1. Cheesy material in bifurcation of bronchi.
Virus isolates from chicken at 4 and 8 days after inoculation of isolate (virus in all four groups) on the other hand, no virus detected in chicken from negative control group. Antibodies serum were detected only in the inoculated chicken at 8 days later which reveal that virus infection induced an immune response in the inoculated birds.
Cross-neutralization assay in chicken embryo
For performing this test Antisera were prepared using SPF chickens from four isolates (X1, X2, X3, and X4) IBV. Antisera titer was determined by ELISA, neutralization of four antisera to homologous strains were in the range 5.5-5.8, referring that each IBV isolate was effectively neutralized by antiserum of homologous strain (Tables 3-5). The neutralization of four antisera to heterologous IBV isolates was in the range of 3.0-4.4, indicating the presence of cross reactive antigens among four IBV isolates. Neutralization of four antisera to the classic IBV strain H120, M41, Conn and Gray were in the range of 8.0 to 1.4 indicating the antigenic differences between four IBV isolates and the classic IBV strains. Furthermore, indicating that antiserum to vaccine strain H120 was unable to effectively neutralize seven IBV isolates. So these steps revealed that serotypes of the four isolate were different from the classic IBV strains H120, M41, Conn and Gray.
| IBV strain | Year of isolation | Serotype/organ used for virus isolation | Geographic origin | Accession no. |
|---|---|---|---|---|
| IBV | 1984 | Unknown | USA | M95169.1 |
| Beaudette CK | 2001 | Strain Beaudette CK | USA | AJ311317.1 |
| IBV-EP3 | 2005 | Chicken embryo cell passage | Singapore | DQ001338.1 |
| IBV-p65 | 2005 | Vero cell | Singapore | DQ001339.1 |
| 1b | 2004 | Beaudette-Vero cell | USA | AY692454.1 |
| ck/CH/LHL | 2010 | Chicken | China | JF828980.1 |
| M41 | 2006 | Massachusett | USA | DQ834384.1 |
| Ind-TN92 | 2003 | Kidneys | India | KR902510.1 |
| ck/CH | 2007 | Unknown | China | JF274479.2 |
| IBV | 2013 | Unknown | China | KF696629.1 |
| IBV | 2012 | Unknown | china | KJ425494.1 |
| H120 | 1960 | Gallus | Netherlands | FJ888351.1 |
| Gray | 1962 | Gray serotype | USA | L14069.1 |
| Conn | 1956 | Connecticut | USA | L18990 |
Table 3: IBVstrains published in GenBank.
| Group | Disease chickens | Dead chickensA | AntibodyB | Reisolated virusC | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MK562377 | 10 | 2/10 | 0/10 | 8/10 | 8/9 | 9/9 | + | + | - | - |
| MK562376 | 10 | 1/10 | 0/10 | 8/10 | 9/10 | 9/6 | + | + | - | - |
| MK562375 | 10 | 3/10 | 0/10 | 9/10 | 9/10 | 8/8 | + | + | - | - |
| MK562374 | 10 | 2/10 | 0/10 | 9/10 | 9/10 | 9/10 | + | + | - | - |
| Control | 0 | 0/10 | 0/10 | 0 | 0 | 0 | - | - | - | - |
Table 4: Experimental infection of chickens with four IBV strains.
| NI | IBV strains | Gray | MK562377 | MK562376 | MK562374 | MK562374 | H120 | Conn |
|---|---|---|---|---|---|---|---|---|
| MK562377 | 5.8 | 4.2 | 3.4 | 3.2 | 0.8 | 0.8 | 1.4 | 0.9 |
| MK562376 | 4.0 | 5.6 | 3.2 | 3.0 | 1.0 | 0.8 | 1.4 | 0.9 |
| MK562375 | 4.2 | 3.4 | 3.2 | 3.0 | 1.0 | 0.8 | 1.4 | 1.0 |
| MK562374 | 4.4 | 4.0 | 5.8 | 5.5 | 0.8 | 0.8 | 1.4 | 1.0 |
| H120 | 1.2 | 1.2 | 1.0 | 1.2 | 4.4 | -- | -- | -- |
| Conn | 1.2 | 1.2 | 1.0 | 1.2 | -- | 4.4 | -- | -- |
| M41 | 1.2 | 1.2 | 1.0 | 1.2 | -- | -- | 4.0 | -- |
| Gray | 1.2 | 1.2 | 1.0 | 1.2 | -- | -- | -- | 4.6 |
Table 5: Result of cross-neutralization tests of different IBV strains.
Phylogenetic analysis
Phylogenetic analysis was performed on the S1 gene for the four isolates and 14 published IB strains. IBV strains were clustered in to 3 group I, II and III as shown in Figure 2. Group I and II comprise of classical vaccine strains whereas, our four strains clustered in the group III revealing antigenic distant and other countries S1 gene.
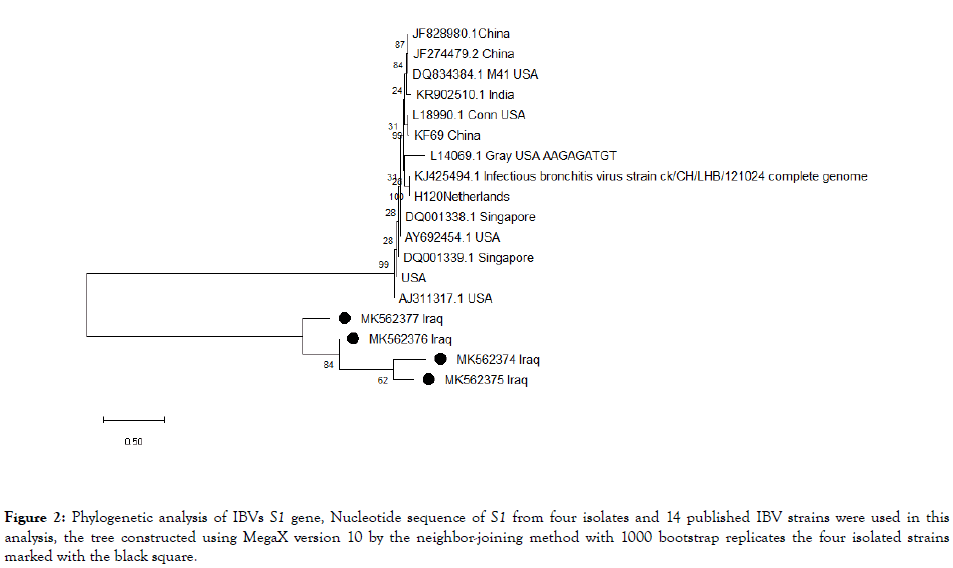
Figure 2. Phylogenetic analysis of IBVs S1 gene, Nucleotide sequence of S1 from four isolates and 14 published IBV strains were used in this analysis, the tree constructed using MegaX version 10 by the neighbor-joining method with 1000 bootstrap replicates the four isolated strains marked with the black square.
Mk562377 were found that sharing some similarities with the Aj311317.1 USA strains and other Iraqi strains had some difference confirming of newly strain with antigenic variation circulating in Iraq.
One of the major diseases in poultry industry is IB. Occurrence of this disease had been a record of severe economic losses in Iraq since it was infected vaccinated and non-vaccinated flock [1,7,11-13]. Inadequate protection against multisera of IB resulting in emergence of new variant as the vaccine program of IB will not protect from the newly strain one which could lead to the outbreak because of, the different serological of new strain with the vaccine strain. Controlling on the IB strains (status) that circulating in the country will help in managing and using appropriate strains which could provide an immune protection since the vaccinated strains known not related to the our strain.
Different chicken farms (different province) were included in this study for the collection of the four strains from dead or disease chickens that have been vaccinated with live vaccine of Massachusetts serotype (H120 and H52) in Iraq during 2007-2019. All the pathogenic characteristics of the IBV were addressed for the four strains in inducing an immune response.
Using antisera against the four IBV strain as well as for the classic strains such as (H120, M41, Conn, and Gray in cross neutralization test in determine the antigenic relationship between the classic strains and our four IBV strains. The result for the four IBV strains was antigenically distinct from classic IBVstrains. Moreover, cross neutralization test revealed that, S1 gene was related IBV serotype but it is not enough in determining the serotype. In this studies of relations ship between cleavage site motifs which is essential for getting in to (cleaving) S1 and S2 subunit during viral mutation [4]. Cleavage site motifs, geographical origin, pathogenicity and serotype relationship have been reported by research [11]. Cleavage sit analysis for the four isolated IBV strains appeared that two of the four strains had the motif (HRRRR) comparing it to the general pattern of R-R-X-R-R was different as it (X amino acid). Motif found that is non-Iraqi strain but it is investigated in other strain in china [1]. There was studies mentioned that the cleavage sit had no relationship with the pathogenicity, tissue tropism [14]. Because of the respiratory and nephropathogenic strains had the same cleavage motifs resulting that cannot consider it as a determinate for the pathotypes.
Phylogenic analysis of the our Four Isolated were clustered in one group III were genetically distant from genotype I and II which included classical vaccine strains as shown in the Figure 1.
This study revealed that, the relationship of the four strains and other published result in the Genbank database were Aj311317.1 USA and USA strain genotype circulating in vaccine and nonvaccine flock in the recent year in Iraq [13] whereas, the other strains were distributed far from our isolate like china, India, M41, Netherland and Singapore.
So this study confirmed that different genotype of IBV were circulating in the population in Iraq. The majority of Iraqi strains belong to the group III and there serotype still need to be identified. From the perspective analysis vaccination do not provide any protection against newly emerge strains, So developing alternative vaccine should be consider for the circulating strains in Iraq in order to effectively control IBV in Iraq.
Citation: Abady NR (2019) Comparative Study of Phylogenetic Analysis of S1 Gene of Infectious Bronchitis Virus Isolates from Iraq with the Selected Strains (genebank). J Antivir Antiretrovir. 11:183. doi: 10.35248/1948-5964.19.11.183
Received: 10-May-2019 Accepted: 17-May-2019 Published: 24-May-2019 , DOI: 10.35248/1948-5964.19.11.183
Copyright: © 2019 Abady NR, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.