Fungal Genomics & Biology
Open Access
ISSN: 2165-8056
ISSN: 2165-8056
Research Article - (2020)
Phytophthora infestans has seven genes coding for glucokinases, with the [PITG_06016] (PiGlcK-1) gene being the highest expression at different stages of growth. However, its role in the metabolism of P. infestans is still unclear. In this study we cloned, overexpressed, purified and biochemically characterized this enzyme. The purified recombinant protein has a Km of 0.66 mM and 2.2 mM for glucose and ATP, respectively. PiGlcK-1 is capable of phosphorylating fructose, although only 48% with respect to glucose, using ADP and pyrophosphate (PPi) as a phosphoryl group donor. These molecules are 50% and 11% less effective than ATP, respectively. In addition, PiGlcK-1 can be activated by PPi increasing its activity 3.6 or 2.5 times when the phosphoryl donor is ATP or ADP, respectively. PiGlcK-1 can form oligomeric complexes and these tend to spontaneously dissociate, with the monomer having the highest activity. This oligomerization can be destabilized by dithiothreitol which increase PiGlcK-1 activity twice. These results reveal that PiGlcK-1 is a versatile enzyme that could play an important role in the use of available sugars during the first stage of infection, as well as in the necrotrophic stage, and suggest a possible regulatory mechanism against possible conditions of oxidative stress.
Phytophthora infestans; Glucokinase; Recombinant protein; Oligomeric complexes; Oxidative stress
Phytophthora infestans is an oomycete that causes the disease called late blight, which affects tomato and potato crops worldwide, causing significant economic losses [1]. Attempts to control this disease include the use of fungicides that are not very effective for the elimination of the phytopathogen and are toxic to humans [2]. Despite having studied P. infestans for more than a century to date, it has not been possible to control the disease caused by this phytopathogen, positioning it as a threat to world food security [3]. A detailed characterization of the metabolism of P. infestans could contribute to the search of solutions against this threat.
Currently, most studies of P. infestans metabolism have been inferred based on mRNA and indicate that glycolysis plays an important role in the infection process levels [4-7]. However, this pathway has been little studied in this organism at enzymatic level. In common with most eukaryotic organisms, hexokinase (EC 2.7.1.1) is the first enzyme of the glycolytic pathway [8]. However, in P. infestans and other stramenopiles, with the exception of Blastocystis hominis as well as other unicellular eukaryotes, such as Trichomonas vaginalis and Giardia intestinalis, this enzyme has been replaced by a glucokinase (EC 2.7.1.2) [6].
Enzymes that catalyze the phosphorylation of D-Glucose and other hexoses through donors of phosphoryl groups such as ATP, ADP, PPi and inorganic Poly-Phosphoryl [Poly (Pi)] have been classified based on their primary structures within two major non-homologous families: the Hexokinase family and the Ribokinase family [9]. The first family is subdivided into three groups: HK, A and B. The HK group harbours hexokinases belonging to eukaryotic organisms that has ATP as a donor of the phosphoryl group, different molecular weights, subcellular distribution and isoenzymes that differ in their kinetic and regulatory characteristics [9,10]. Group A is represented by enzymes belonging to Gram-negative bacteria, cyanobacteria, amytochondriate protists and trypanosomatids with molecular weights between 33 to 42 kDa. Finally, group B is represented by enzymes belonging to Gram-positive bacteria and crenarqueota organisms. These last two groups comprise enzymes known as Glucokinases (GlcKs) capable of exclusively phosphorylating Dglucose [9-11]. On the other hand, the Ribokinase family comprises enzymes that only use ADP as a donor of the phosphoryl group and have molecular weights between 47 to 54 kDa [9,12].
P. infestans presents up to seven genes for GlcKs [13] which are differentially transcribed both in nutritional media and in the different life stages of P. infestans [6,14,15], which denotes the complexity and adaptability of this organism. One of these glucokinases, [PITG_06016] available in the National Centre for Biotechnological Information (NCBI) database (http:// www.ncbi.nlm.nih.gov) (NCBI database) that we will call PiGlcK-1 here, is more expressed in zoospores, germinated cysts and appressorium compared to the other P. infestans glucokinases [15], showing eight times greater expression in zoospores than in hyphae [14,15]. The presence of GlcKs in this organism instead of hexokinases could restrict the use of other equally abundant sugars that are present in plants [6] which are usually mobilized using membrane-bound transporters in hyphae or haustoria [16]. In this sense, most of the genes expressed during the development of the haustoria encode for a transporter of sugars (HTX1), specialized for D-glucose and Dfructose, confirming that these sugars are the primary carbohydrates imported into the haustoria [17]. Only a few species, especially bacteria, are known to contain true glucokinases [10]. Therefore, it is important to know if the P. infestans GlcKs are true glucokinases or if they are able to use other important sugar in plants.
With the intention of understanding more about the metabolism of P. infestans, we have carried out in this study, a biochemical characterization of PiGlcK-1. We describe the kinetic properties of the recombinant enzyme, the effect of dithiothreitol (DTT) on PiGlcK-1 activity, as well as its oligomeric states. We propose a metabolic model where PiGlcK-1 activity can be regulated under conditions of oxidative stress during the progress of P. infestans infection.
Cloning of PiGlcK-1 gene
The PiGlcK-1 gen [PITG_06016] retrieved from NCBI database (http://www.ncbi.nlm.nih.gov) was amplified by PCR from genomic DNA obtained from P. infestans, isolate SR 1.8-04 (homothallic, originally isolated from potato var. Granola, grown at Santa Rosa, Mérida, Venezuela) [18]. The forward primer was 5´-GCATATGTCTGACTACAGCCTCATCATC-3 ´ (NdeI site in bold) and the reverse primer was 5 ´-CGGATCCCTACGCGCTCGAGTGCAAG-3´ (BamHI site in bold). The amplification mixture (50 ml) contained 1 mg of P. infestans genomic DNA, 1 mM of each primer, 200 mM of each of four deoxynucleotides, 1.5 mM MgCl2 and 1.5 U of DNA polymerase (DNA polymerase Platinum, Invitrogen) with the corresponding PCR buffer. PCR for glucokinase gene was performed as follows: an initial incubation at 95oC for 2 min, 30 cycles of denaturation at 95oC for 2 min, annealing at 55oC for 45’s, and extension at 70oC for 90’s; the final incubation was at 70oC for 10 min.
The PCR products were ligated into the pGEM-T vector (Promega) cloned and sequenced using the T7 and SP6 primers in an automated sequencer. The gen was transferred to the pET28a vector (Novagen) and used to transform the Escherichia coli strain BL21pRARE for the production of the recombinant PiGlcK-1 with a 20 residue-long N-terminal containing a His-tag.
Multiple sequence alignments and phylogenetic analysis
Sequence data were obtained from the NCBI, which selected sequences homologous to the PiGlcK-1 protein. Multiple alignments were performed using MUSCLE [19] with predetermined parameters. Protein sequences similarity analysis was performed with the ESPript 3.0 software (http:// espript.ibcp.fr/ESPript/ESPript/).
The sequences for the phylogenetic analysis were obtained as described in the previous section. The alignment was performed with MUSCLE [19] and a subsequent refinement with TCS [20]. To perform the tree, the substitution model that best fit the data was previously evaluated using the Model Finder program [21], with LG+G4 being the best model. The tree was generated by PhyML with the chosen substitution model, with 500 bootstrap non-parametric replicas, the gamma estimate and the optimized distribution parameter of BIONJ. The tree was edited with ITOL v5 [22]. The access numbers of the sequences used were added to the tree legend.
Overexpression of PiGlcK-1 in Escherichia coli and their purification
Cells were grown at 25oC for 48 h in ZYM-5052 auto induction medium (0.5% (w/v) yeast extract, 25 mM Na2HPO4, 25 mM KH2PO4, 50 mM NH4Cl, 5 mM Na2SO4, 2 mM MgSO4, 0.5% (w/v) glycerol, 0.05% (w/v) glucose and 0.2% (w/v) lactose) [23] supplemented with 50 ml/ml kanamycin and 100 ml/ml chloramphenicol, to O.D 600 nm values of 10. Cells from 50 ml culture were harvested by centrifugation, washed with PBS, and resuspended in 10 ml lysis buffer (100 mM HEPES pH 7.6 and 0.1% Triton X-100) in the presence of a cocktail of protease inhibitors (Sigma). The resulting suspension was broken by sonication and the lysate centrifugated at 12,000 g for 15 min at 4oC. The supernatant contained recombinant His-tagged glucokinase.
PiGlcK-1 was purified using a HisLink column (Amersham Biosciences). The column (2 × 0.8 cm) was equilibrated with the PBS and washing with the same buffer, His-PiGlcK-1 was eluted with 200 mM imidazole. The imidazole was removed from the sample using a PD-10 column (Amersham Biosciences). The purity of the PiGlcK-1 was checked by SDS-PAGE followed by Coomassie blue R-250 staining.
Antibodies
A polyclonal antiserum against purified recombinant PiGlcK-1 was raised in a rabbit by intradermic injection with 0.2 mg of purified protein in incomplete Freund`s adjuvant (1:1) at multiple sites. The immune response of the rabbit was boosted five times, at two-week intervals, using the same conditions as the first injection, except for the last boost which was performed in the absence of the adjuvant. The rabbit was bled by cardiac puncture two weeks after the four immunizations. The sera were stored at 20°C.
Enzyme assays
Glucokinase activity was assayed by measuring the formation of NADH by coupling the phosphorylation of D-glucose to the reduction of NAD+ by glucose-6-phosphate dehydrogenase (G6PDH), measured at 340 nm and 25oC using a Hewlett- Packard 8452 diode array spectrophotometer. For the assay of Dfructose phosphorylation, phosphoglucose isomerase (PGI) was added to the reaction mixture instead G6PDH. The assay was performed in a 1-ml cuvette containing 100 mM triethanolamine (TEA), pH 7.6, 5 mM MgCl2, 5 mM ATP, 4 mM D-glucose or Dfructose, 0.72 mM NAD+, 10 mM DTT in the presence of purified PiGlcK-1 and 1 U.ml−1 G6PDH (type XXIV from Leuconostoc mesenteroides, Sigma) for glucose or 1 U.ml−1 PGI (Saccharomyces cerevisiae, Sigma) for fructose [24]. The Km for glucose was determined by varying its concentration between 0.1 mM and 0.50 mM, whereas the Km for ATP was measured with concentrations varying between 0.5 and 5 mM. The effect of DTT on the glucokinase activity was determined by varying its concentration between 0 and 50 mM. The glucokinase activity with 5 mM ADP or 5 mM PPi (in absence of ATP) and the effect of 1 mM PPi on the glucokinase activity (in presence of ATP or ADP) were determined similarly to glucose assay described above. Values of kinetic parameters were calculated by plotting the data and curve fitting using the appropriate equations in the program SigmaPlot 11.0 SPSS. One unit (U) of enzyme activity is defined as the amount of PiGlcK-1 that catalyzes the formation of 1 mmol of product per minute or consumption of 1 mmol substrate per minute.
The effect of hydrogen ion concentrations on PiGlcK-1 activity was measured using a polybuffer of MES, MOPS, Tris and TEA, each at 25 mM, over a pH range from 7 to 9.5. At each pH condition at which the activity was assayed, the time-dependent absorbance variation at 340 nm was recorded until it reached the steady state.
For the preparation of the b-D-glucose anomer, 10 mM of α-Dglucose (Sigma) was dissolved in cold water (4°C), which was heated for 10 min at 60°C and kept on ice to shift the anomeric equilibrium towards the formation of β-D-glucose [25].
Protein determination, SDS-PAGE and immunoblotting
Proteins were quantitatively assayed by the Bradford method [26] with bovine serum albumin as standard. SDS-PAGE was performed in 12.5% polyacrylamide gels according to the method described by Laemmli [27]. Proteins separated by SDSPAGE were transferred to a nitrocellulose membrane (Amersham Bioscience) and Western blotting was performed as described [28]. The nitrocellulose membrane (Thermo Scientific) was first blocked with PBS containing 10% casein and 0.1% Tween 20 for 2 h. After washing with PBS containing 0.1% Tween 20, the membrane was incubated with the anti- PiGlcK-1 polyclonal obtained in rabbit (1:1000 dilution) for 1 h, then incubated with a rabbit anti-IgG conjugated with alkaline phosphatase (dilution 1:5,000) for 1 h and visualized with BCIPNBT in PBS.
Cloning and sequence analysis of the P. infestans glucokinase PiGlcK-1
PiGlcK-1 gene of P. infestans was identified in the NCBI database [PITG_06016] and amplified by PCR using genomic DNA from P. infestans and cloned. It encodes a polypeptide of 353 amino acids with a calculated molecular mass and isoelectric point of 38 kDa and 5.2, respectively. A comparison of PiGlcK-1 with homologous sequences identified by MUSCLE [19], showed the following degrees of identity with group-A GlcKs from other stramenopiles organisms: Phytophthora parasitica (90.4%), Plasmopara halstedii (79.4%) and Phaeodactylum tricornutum (38.6%). A low identity was also found with a counterpart in the prokaryote E. coli (32.1%) and Pseudomonas aeruginosa (28.4%) (Figure 1).

Figure 1: Sequence alignment of glucokinase-1 (PiGlcK-1) from Phytophthora infestans [PITG_06016] with representative GlcKs from others species. Escherichia coli (AAA64506.1), Pseudomonas aeruginosa (NP_251883.1), Phaeodactylum tricornutum (XP_002183440.1), Plasmopara halstedii (XP_024577148.1), Phytophthora parasitica (ETM99186.1). Squares: amino acids involved in glucose binding, black circles: amino acids involved in ATP binding, triangle: catalytic aspartate, asterisks: cysteine residues present in the PiGlcK-1 sequence of P. infestans.
Further analysis of the deduced amino acid sequence showed that PiGlcK-1 contains the characteristic ATP binding site 8I-XX- D-X-G-G-X-N-X-R-X-X-L21 (Asp, Gly, Asn, Arg and Leu in bold) and conserved glucose binding residues (Asn101, Glu159, His162 and Glu189) present for GlcK from E. coli [29]. The above indicates that PiGlcK-1 belongs to group A of the Hexokinase family. Interestingly, we also observed that the PiGlcK-1 sequence shows ten cysteines, with Cys12 within the ATP binding motif and Cys172 and Cys175 near residues involved in glucose binding (Figure 1).
Phylogenetic analysis
Figure 2 shows the phylogenetic analysis obtained with PhyML for 80 Glck sequences of P. infestans and other organisms. PiGlck-1 branches with those from other species of Phytophthora and other oomycetes in the well supported stramenopiles clade, clearly differentiating it from other eukaryotes. The amino acid identity in this clade is more than 90% in contrast to 50% or less in other groups of bacteria, cyanobacteria and fungi.
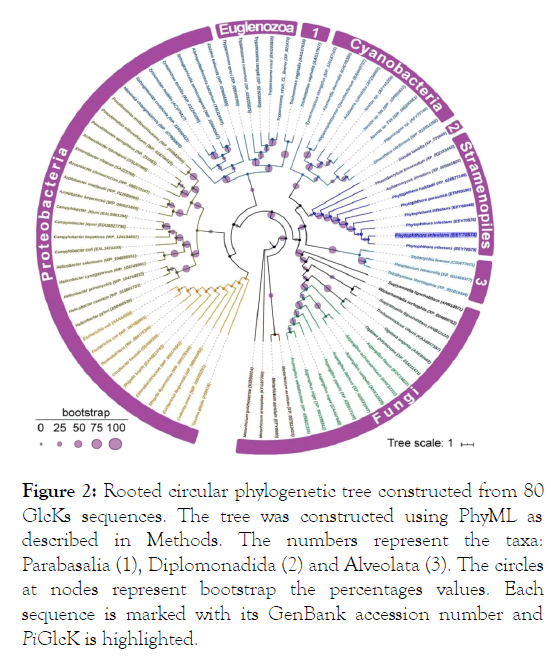
Figure 2: Rooted circular phylogenetic tree constructed from 80 GlcKs sequences. The tree was constructed using PhyML as described in Methods. The numbers represent the taxa: Parabasalia (1), Diplomonadida (2) and Alveolata (3). The circles at nodes represent bootstrap the percentages values. Each sequence is marked with its GenBank accession number and PiGlcK is highlighted.
Expression of recombinant PiGlcK-1 in E. coli and purification of the protein
The complete gene for PiGlcK-1 was expressed in E. coli strain BL21pRARE in order to investigate their biochemical and kinetic properties. The coding sequences were ligated to the expression vector pET28a to produce a fusion protein with an amino terminal (His)6-tag. Bacteria were grown in autoinduction medium and a high percentage of insoluble enzyme was obtained. However, this expression system allowed us to produce enough amounts of soluble recombinant protein (Figure 3A). After induction, the soluble fraction was purified to homogeneity by HisLink column, using 200 mM imidazole concentration for the elusion (Figure 3B). The purified protein was used to raise antisera, and for its respective kinetic characterization.
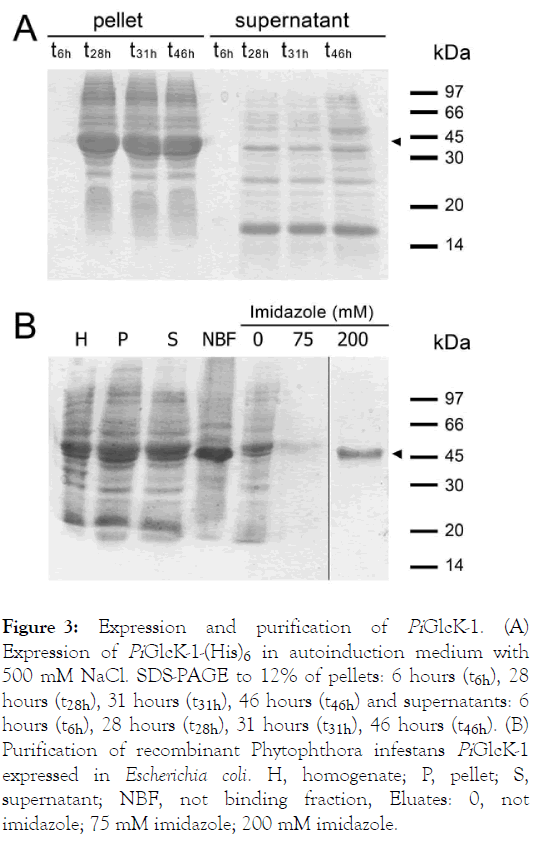
Figure 3: Expression and purification of PiGlcK-1. (A) Expression of PiGlcK-1-(His)6 in autoinduction medium with 500 mM NaCl. SDS-PAGE to 12% of pellets: 6 hours (t6h), 28 hours (t28h), 31 hours (t31h), 46 hours (t46h) and supernatants: 6 hours (t6h), 28 hours (t28h), 31 hours (t31h), 46 hours (t46h). (B) Purification of recombinant Phytophthora infestans PiGlcK-1 expressed in Escherichia coli. H, homogenate; P, pellet; S, supernatant; NBF, not binding fraction, Eluates: 0, not imidazole; 75 mM imidazole; 200 mM imidazole.
Kinetic characterization of recombinant PiGlcK-1
Recombinant PiGlcK-1 was characterized with respect to its kinetic properties. The obtained progress curve showed a delay (lag) of several minutes until reaching the steady-state velocity, which was used to determine all the kinetic parameters. PiGlcK-1 showed hyperbolic kinetics for glucose and ATP, indicating classic Michaelis-Menten behavior. The Km values for glucose and ATP were determined according to the values obtained from the Lineweaver-Burk graph, being 0.66 mM and 2.2 mM, respectively (Figure 4).
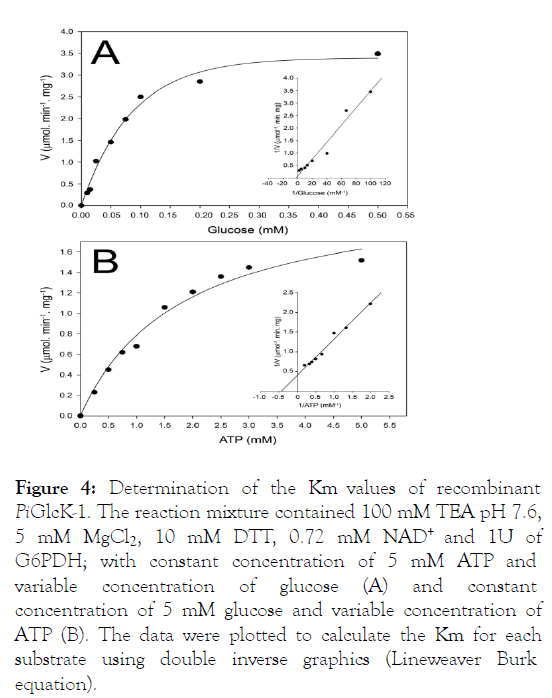
Figure 4: Determination of the Km values of recombinant PiGlcK-1. The reaction mixture contained 100 mM TEA pH 7.6, 5 mM MgCl2, 10 mM DTT, 0.72 mM NAD+ and 1U of G6PDH; with constant concentration of 5 mM ATP and variable concentration of glucose (A) and constant concentration of 5 mM glucose and variable concentration of ATP (B). The data were plotted to calculate the Km for each substrate using double inverse graphics (Lineweaver Burk equation).
This Km value for glucose is very similar to those reported by other GlcKs from E. coli (0.78 mM), Bacillus stearothermophilus (0.52 mM) or T. cruzi (0.94 mM) [10,11,30]. Interestingly PiGlcK-1 can also use the α-D-glucose (14 U.mg-1) and β-DGlucose (9.8 U.mg-1) anomers. To determine if PiGlcK-1 was a true glucokinase, we assessed the enzyme's ability to phosphorylate fructose, checking that PiGlcK-1 can phosphorylate this sugar by 48% relative to glucose (Table 1).
The enzymes of the large Hexokinase family (groups HK, A or B) catalyse the phosphorylation of hexoses and in some cases only glucose by means of phosphoryl donors (ATP, ADP, PPi and inorganic polyphosphate [poly (P)] [9,10,31-33]. To analyze whether PiGlcK-1 could use other phosphoryl group donors, various enzyme assays were performed in the presence of ADP or PPi. We were able to determine that PiGlcK-1, like other GlcKs, particularly those from Archaea, can use ATP and ADP. However, ADP was less efficient, that is to say around 50%, concerning ATP. Similarly, a small activity of 11% with respect to ATP was found, when the phosphoryl donor was PPi (Table 1). These results are similar to that of other GlcKs, particularly in GlcKs from Archaea or B. stearothermophilus, which can use both ATP and ITP as nucleotide substrate [10]. Even the GlcK of the hyperthermophilic archon Pyrococcus furiosus uses ADP as a phosphoryl donor and has no detectable activity with ATP or other potential donors such as GDP or pyrophosphate [34]. Another unusual example is provided by GlcK from the anaerobic bacterium Propionibacterium shermanii, in which this enzyme can use ATP as a phosphoryl donor. However, it is much more specific for polyphosphates [35].
| Molecules | U.mg-1 | % Activity |
|---|---|---|
| Sugars | ||
| α-D-Glucose (4 mM) | 14 | 100 |
| β-D-Glucose (4 mM) | 9.8 | 70 |
| D-Fructose (4 mM) | 6.7 | 48 |
| Donator of phosphoryl group | ||
| ATP (5 mM) | 14 | 100 |
| ADP (5 mM) | 7 | 50 |
| PPi (5 mM) | 5.3 | 11 |
| ATP (5 mM)+PPi (1 mM) | 49.7 | 357 |
| ADP (5 mM)+PPi (1 mM) | 35 | 250 |
All assays contained 10 mM DTT
Table 1: PiGlcK-1 activity with different substrates
Pyrophosphate has been previously reported to serve as an inhibitor of the activity of some kinases [36-39]. For this reason, we tested PPi as a possible inhibitor of PiGlcK-1 activity in the presence of ATP or ADP. Surprisingly, in the presence of PPi, PiGlcK-1 activity, far from being inhibited, showed an increase of more than three times when the phosphoryl donor was ATP and more than two times when the phosphoryl donor was ADP (Table 1). A study of ATP-dependent PiGlcK-1 activity by varying PPi concentrations showed that it peaked at a PPi concentration between 0.75 and 1 mM. Interestingly, this activation decreased when PPi concentrations were greater than 1 mM (Figure S1). Until now, there are no reports of other eukaryotic GlcKs that are activated by PPi, and when it has exerted some effect it has been as an inhibitor [10].
The effect of the hydrogenation concentration on the PiGlcK-1 activity showed an optimal value for pH between 9.0 and 9.5 (Figure S2). This optimal pH is similar to those obtained for GlcKs from different organisms [11,40,41].
Effect of Dithiothreitol on the oligomeric forms and the activity of PiGlcK-1
Glucokinases are characterized by a high susceptibility to the presence of reducing agents such as DTT or glutathione [42-46]. In our case, the activity of PiGlcK-1 showed an increase (more than double the activity) in the presence of 10 mM DTT. However, higher concentrations of DTT produced an inhibitory effect on its activity (Figure 5A). The effect of ionic strength was ruled out since the activity of PiGlcK-1 in the presence of other salts did not show an increase in the activity of the enzyme. This phenomenon of DTT activation has been reported for several kinases from various organisms [40,47-50]. Interestingly, an analysis of the PiGlcK-1 sequence showed the presence of ten cysteines that could potentially participate in the formation of multiple inter and intramolecular disulfide bonds, which would be sensitive to reducing agents.
Figure 5: Effect of Dithiothreitol on the oligomeric forms and activity of PiGlcK-1. (A) Effect of the DTT concentration on the activity. The enzyme activity was measured as described under materials and methods. The reaction mixture contained 100 mM TEA pH 7.6, 5 mM MgCl2, 5 mM glucose, 5 mM ATP, 0.72 mM NAD+ and 1U of G6PDH and variable concentration of DTT (0-50 mM). (B) Western immunoblotting analysis after denaturalizing electrophoresis using anti-PiGlcK-1. 30 μg of PiGlcK-1 purified was incubated with 0 mM Dithiothreitol (DTT), 10 mM and 50 mM.
In order to see if the effect of DTT on PiGlcK-1 could also be related to the presence of possible oligomeric states of the enzyme, a Western blot was analyzed in the presence and absence of DTT. It was observed that PiGlcK-1 in the absence of DTT can be found in different oligomeric states (Figure 5B). Although several bands were observed, the most evident corresponds to the monomer (38 kDa) in addition to lower intensity bands corresponding to a dimer (90 kDa) and oligomeric states with masses greater than the latter. Interestingly, the signal from the high molecular weight bands decreased in intensity when the DTT concentration was increased from 10 mM to 50 mM. This result could suggest the presence of oligomeric forms (involving disulfide bridges), in addition to the monomeric form of the enzyme. Interestingly, a double band electrophoretic pattern was observed in the region corresponding to the monomer, both under reducing and nonreducing conditions. This behavior has already been observed for human glucokinase and can be interpreted as an incomplete reduction of disulide bonds to which DTT would not have had access due to the spatial arrangement of the residues of cysteine in protein [51]. Therefore, this incomplete reduction could give rise to two populations of monomeric PiGlcK-1 whose conformational states can be evidenced electrophoretically.
In this study we cloned, overexpressed, and kinetically characterized the P. infestans PiGlcK-1 [PITG_06016]. Based on its sequence, this enzyme belongs to the glucokinase group A from the Hexokinase Family by code IPR003836 [9,52]. However, our kinetic studies showed that PiGlcK-1 can phosphorylate both glucose and fructose, indicating that it is not a true glucokinase. Another unusual case for group A enzyme that are not specific for glucose is the GlcK from E. coli B [40]. Furthermore, the evidence that PiGlcK-1 can use ATP, ADP or PPi as donors of phosphoryl groups, suggests that its current classification as a strict glucokinase should be reviewed and this suggests that group A from Hexokinase family can contain a more varied type of enzymes.
Surprisingly, when we included PPi as possible inhibitors of PiGlcK-1 activity in the presence of ATP or ADP, the enzyme was activated 3.6 times and 2.5 times when the phosphoryl donor was ATP or ADP, respectively. These results suggest the presence of a PPi binding site different from that of ATP, as reported for Zymomonas mobilis GlcK, which is activated by phosphate [53]. These metabolites could be related with to a low energetic status in cell, contributing in the regulation of this enzyme. In addition, it has already been reported that oomycetes diverge from the classic form of eukaryotic glycolysis by expressing phosphofructokinase (PFK) that use PPi (EC 2.7.1.90) instead of ATP (EC 2.7.1.11) as a phosphoryl donor [4]. Thus, although several ancestors of oomycete glycolytic enzymes are assumed, PiGlcK-1 forms a solid clade, along with other species of Phytophthora and other oomycetes, reflected by the high identity in their amino acid sequence, perhaps due to the particularity of using ATP, ADP and PPi as phosphoryl donors.
Another interesting fact was the activation of PiGlcK-1 by the reducing agent DTT. This and other sulfhydryl compounds play a role in the stability and catalytic activity of GlcK in various organisms [42-48]. Interestingly, PiGlcK-1 showed ten cysteines in its sequence, of which three are close to the glucose and ATP binding motif (Cys12, Cys172 and Cys175) and could possibly play a role in enzymatic activation. The other cysteines may be involved in oligomeric associations mediated by intermolecular disulfide bridges.
The reduction of disulfide bonds affects the molecular flexibility of proteins [51], which may favours the destabilization of possible oligomeric states. Here we obtained electrophoretic evidence indicating that in the presence or absence of DTT, PiGlcK-1 can be found in different oligomeric states, whose transition would be associated to a greater or lesser activity of the enzyme and that, could possibly be related to the delay of this activity observed in the progress curve. This aggregation behaviour involving cysteines has been reported for cytosolic glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from Arabidopsis thaliana, which under conditions of oxidative stress forms inactive aggregation states, which can be reversed in the presence of DTT [54]. It has also been proposed that these disulfide bonds could fulfil the role of redox switches rather than as stabilizers of the native structure [55]. In the case of PiGlcK-1, the susceptibility to the reduction of disulfide bonds, under certain extracellular environments of high oxidative stress, could represent a sensor of such changes, allowing this enzyme to modulate its activity.
In accordance with this, it has previously been reported that in solanaceous plants, infection by P. infestans involves the release of Reactive Oxygen Species (ROS) [56]. In response to this, there is a large abundance of enzymes related to redox metabolism in germinated cysts and P. infestans appressorium to combat oxidative stress [15]. Therefore, the high expression of PiGlcK-1 in structures related to the establishment of infection could suggest a relationship with high levels of PiGlcK-1 and the need for a high supply of NADPH to fuel this redox metabolism. Thus, PiGlck-1 would not only be limited to energy generation through glycolysis, phosphorylating available D-glucose and Dfructose, but would also provide glucose-6-phosphate necessary to feed the pentose-phosphate pathway generating NADPH. Actually, PiGlcK-1 can phosphorylate both a- and b-glucose, giving rise to a- and b-glucose-6-phosphate (G6P), respectively. In this sense, the anomeric selectivity of G6PDH by b-G6P [57-59] would improve the metabolic flow towards the pentose phosphate pathway. Moreover, the consumption of b-G6P by G6PDH, together with a spontaneous anomerization of the alpha to beta form, would further contribute to the displacement of this balance, decreasing the concentration of a G6P. This would favour fructose phosphorylation to feed the glycolytic pathway.
The role of various GlcKs that are expressed at all stages of the life cycle of P. infestans is still unclear, but differences regarding the specificity of sugars and phosphoryl donors, as well as a diversification of functions, indicate that a mechanism efficient for feeding during the progress of infection should be considered. In this sense, the fact that PiGlcK-1 is actually behaving as a HK, would allow it to use glucose and fructose present in the apoplast of the host cell during the biotrophic phase of infection [7,15]. Additionally, an invertase activity has been reported during infection, which makes use of the sucrose present in the plant to produce D-glucose and D-fructose that can be transported into the phytopathogen [17,60].
This possible participation of PiGlcK-1 during the establishment of the infection to take advantage of other sugars available at this stage coincides with that observed in the transcriptomic models of the dynamics of tomato infection by P. infestans, where it is observed that the reactions of P. infestans are enriched for glycolysis at the beginning of the infection. However, as the infection progresses, this plant pathogen becomes less dependent on its own metabolism and can continue to obtain its nutrients through the transport of metabolites from the tomato necrotic lesion, including D-fructose [7]. All of the above would allow the maintenance of an optimal glycolytic flow not only for obtaining energy but also for supplying the NADPH necessary for the survival of the pathogen in conditions of oxidative stress as a result of the hypersensitive response of its host.
The characterization of the recombinant enzyme PiGlcK-1 gives us indications of a diversification of functions of this enzyme, as well as of its possible role in the metabolism of P. infestans. In this sense, we propose a versatility of this enzyme to phosphorylate D-glucose and D-fructose present during the first stage of infection. While, during the necrotrophic stage, when ATP concentrations could be lower, PiGlcK-1 would continue to phosphorylate sugars thanks to its ability to use other donors of phosphoryl groups, as well as activation by PPi, ADP or DTT. Furthermore, PiGlcK-1 could respond to variation in a redox environment, modulating its activity and increasing glucose phosphorylation for NADPH synthesis.
Citation: Villalobos-Piña L, Balza H, Acosta H, Andrade-Alviarez D, Rojas A, Avilan L, et al. (2020) Characterization of Glucokinase-1 from Phytophthora infestans: A Versatile Enzyme. Fungal Genom Biol. S1: 001.
Received: 13-Nov-2020 Accepted: 27-Nov-2020 Published: 04-Dec-2020
Copyright: © 2020 Villalobos-Piña L, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.