Translational Medicine
Open Access
ISSN: 2161-1025
ISSN: 2161-1025
Research Article - (2023)Volume 13, Issue 4
Introduction: Cervical cancer is the fourth most prevalent cancer among women worldwide and is a significant contributor to cancer- related deaths, with an estimated 300,000 women losing their lives to the disease annually. Most of these fatalities occur in Low and Middle Income Countries (LMICs), such as Uganda, where access to screening and treatment options is limited. Early detection of cervical cancer is crucial to improve the chances of survival for patients. Currently, cervical cancer screening is typically performed through pap smears, which involve manual examination of cervical samples for abnormalities by medical experts. This process is costly, time-consuming and prone to errors, leading to inaccurate diagnoses. Therefore, it is essential to find more effective and efficient alternative methods for cervical cancer screening to improve access in LMICs and alleviate the burden of cervical cancer.
Objective: The purpose of this study is to develop an automated pre-cervical cancer screening algorithm to detect precancerous cervical lesions.
Materials and methods: We developed a cancer screening algorithm using a 21 layer deep-learning convolution neural network trained on a dataset of 2300 images collected from local sources and some obtained from Kaggle.
Results: The best-performing classifier had an Area Under Curve (AUC) of the accuracy of 91.37%, a precision of 88.80%, a recall of 94.69%, an F1 score of 91.65% and an AUC of 96.0%.
Conclusion: The development and implementation of automated pre-cervical cancer screening algorithms have the potential to revolutionize cervical cancer detection and contribute significantly to reducing the burden of the disease, particularly in resource-limited settings.
Cervical cancer; Deep learning; Screening; Machine learning
Cervical cancer is a preventable and curable disease through screening and vaccination. Despite this, it remains a significant cause of death among women globally. It is the fourth most common form of cancer among women worldwide and results in approximately 300,000 deaths annually [1]. Low and Middle Income Countries (LMICs), such as Uganda, account for almost 90% of these deaths due to limited access to screening and treatment. In response, the World Health Organization (WHO) called for global action to eliminate cervical cancer in May 2018. The WHO developed an ambitious strategy to guide the elimination of cervical cancer as a public health problem, which the World Health Assembly (WHA) adopted in August 2020 [2]. According to this strategy, all countries must maintain an incidence rate of below four per 100,000 women for cervical cancer to be eliminated. To achieve this, the WHO established a 90-70-90 target to be met by 2030. This target involves fully vaccinating 90% of girls with the Human Papilloma Virus (HPV) vaccine by age 15, screening 70% of women aged 35-45 years using a high-performance test and managing 90% of women with pre-cancer and 90% of women with invasive cancer.
The current cervical cancer screening practice in Uganda relies on opportunistic screening using Visual Inspection with Acetic acid (VIA), specifically targeted towards women aged 25-49 at a 3-year interval for Human Immunodeficiency Virus (HIV) negative women and every year for HIV positive women [3]. However, the lack of resources and infrastructure deficiencies have led to low screening participation rates [4]. Another cervical cancer screening method is the Papanicolaou (Pap smear), but this approach is time consuming, labor intensive and therefore less effective in low resource constrained environments [5,6].
Uganda has one of the highest rates of HPV in the world, estimated at 33.6%. Consequently, Uganda also has one of the highest incidence rates of cervical cancer, with about 54.8 cases per 100,000 people annually and a mortality rate of about 41.4 per 100,000 [7,8]. If Uganda is to achieve the WHO target by 2030, more effective early identification of cervical cancer and treatment programs will be required. But these efforts will require adequate financial resources, the development of better screening infrastructure and increasing access to treatment. It is against this background that we propose to develop an Artificial Intelligence (AI) algorithm for automatic cervical cancer image-based diagnosis. Recent advancements in AI, particularly computer vision deep learning techniques, can provide solutions to overcome unsolved colposcopic bottlenecks. AI-based algorithms can learn features of cervical lesions from annotated colposcopy images, which can then be integrated into an automated screening system. We believe that automating the colposcopy examination by creating a cervical cancer screening model will reduce potential false negatives and positives, improving the accuracy of colposcopy diagnosis and cervical biopsy.
The use of deep learning Convolutional Neural Networks (CNN) in automating cervical cancer screening has gained significant attention in recent years due to their ability to effectively analyze and interpret digital images, leading to improved accuracy and efficiency in screening cervical cancer. A number of researchers have developed classification techniques for automated cervical cancer diagnosis using CNNs. In this section, we will discuss some of the most relevant and recent studies in this area, highlighting their methodologies, strengths and limitations.
The study conducted by Sompawong, et al. [9] utilized the Mask Regional Convolutional Neural Network (Mask R-CNN) to automate cervical cancer screening using images obtained from pap smear slides. This study is notable as it was the first to use the Mask R-CNN algorithm to analyze and detect the nucleus of cervical cells, thereby classifying normal and abnormal nuclear features. The proposed algorithm achieved a classification accuracy of 89.8%, with a sensitivity of 72.5% and a specificity of 94.3%.
In their study, Kavitha, et al. [10] utilized ant colony optimization- enabled CNN deep learning technique to extract features and subsequently employed three different algorithms, namely CNN, Multi-Layer Perception (MLP) and artificial neural network, to classify cancerous and non-cancerous cervical images. The CNN classifier showed the highest accuracy among the three algorithms with 95.2%. However, it should be noted that the study used a relatively small dataset, the Herlev dataset and as such, the best-performing model may not be suitable for deployment beyond the academic setting due to potential limitations in generalization caused by the dataset’s size [11]. In another study, Priyanka, et al. [12] utilized the Herlev dataset to design a model using pre-trained ResNet50 network to classify between normal and abnormal cervical cancerous cells. The proposed approach achieved a classification accuracy of 74.04%.
Chandran, et al. [13] put forward two deep learning CNN architectures, namely the VGG19 model and the Colposcopy Ensemble Network (CYENET), to detect cervical cancer using colposcopy images. The study results indicated that the CYENET model outperformed the VGG19 model, achieving higher sensitivity and specificity of 92.4% and 96.2%, respectively. The CYENET model also achieved a classification accuracy that was 19% higher than that of the VGG19 model. Another study conducted by Guo, et al. [14] presented a deep learning algorithm designed to detect in-focus cervical images captured with smartphones. The study compared the performance of three deep learning networks, including retina net (an object detection model), fine-tuned deep learning models (Visual Geometry Group (VGG) and Inception) and transfer learning models (VGG and inception feature extractor+Support Vector Machine (SVM)). These models were evaluated based on their ability to distinguish between sharp and not sharp images. The results showed that the retina net model outperformed its counterparts, although it required more training time to achieve a higher classification performance compared to the second-best model, which was fine- tuned VGG.
Chauhan, et al. [15] conducted a study to assess and analyze the impact of the number of channels in the CNN classification model on the classification of multi-class Liquid-Based Cytology (LBC), Whole Slide Images (WSIs). They proposed a CNN model with two convolutional layers and two pooling layers with different numbers of channels, including (4,8), (8,16) and (32,64). The results revealed that the model with the most channels achieved the best performance across all evaluation metrics, with an accuracy of 96.89%, precision of 93.38%, sensitivity of 93.75% and F-score of 94.15%. Kanavati, et al. [16] conducted a study in which they developed a deep-learning model to classify WSIs of LBC specimens into neoplastic and non-neoplastic categories. Similarly, Cheng, et al. [17] utilized WSIs to train and validate a recurrent neural network algorithm designed to evaluate the degree of lesion present in WSIs.
Alyafeai, et al. [18] proposed a deep learning pipeline that utilized two pre-trained models to automate cervix detection and cervical tumor classification. According to the authors, their proposed classifier outperformed all existing algorithms in terms of classification accuracy and speed. Specifically, the algorithm achieved an Area Under Curve (AUC) score of 0.82 while classifying each cervix region 20 times faster. Due to its accuracy, speed and lightweight architecture, the proposed pipeline is highly suitable for deployment on mobile phones, which is not the case for other reviewed models.
Although several studies have proposed algorithms to detect cervical precancerous cases using various imaging techniques, these algorithms are not readily available to other researchers for deployment. Therefore, this study aims to develop a new algorithm that can be integrated into Uganda’s Electronic Medical Records (EMR) to facilitate easy and efficient screening of cervical precancerous cases among women living with HIV by health workers. By developing a new screening algorithm that is readily available, other researchers and medical professionals will have access to a valuable tool that can improve the accuracy and efficiency of cervical cancer screening programs in Uganda and beyond.
We employed the deep-learning CNN technique to automate cervical cancer screening. Deep learning is a type of machine learning that involves using CNNs to analyze large amounts of data and make predictions or decisions [19]. In detecting cervical cancer, a deep learning algorithm was trained on large datasets of cervical images to identify patterns and features indicative of cancer. The implementation framework of this study was classified into different stages such as collection and annotation of colposcopy images, data preprocessing, model selection, training, validation, testing and deployment of the algorithms. Figure 1 shows the architectural flow diagram of the implementation methodology (Figure 1).
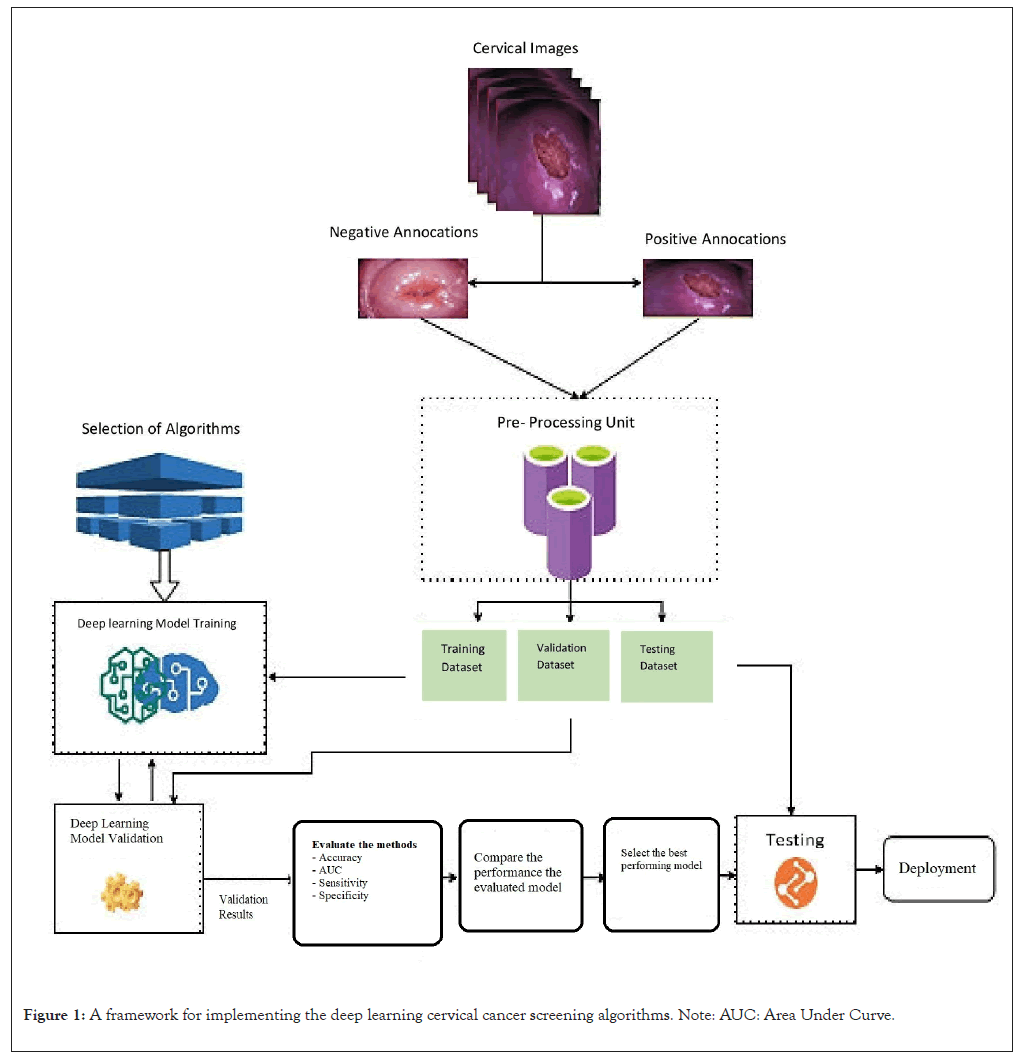
Figure 1: A framework for implementing the deep learning cervical cancer screening algorithms. Note: AUC: Area Under Curve.
Dataset and preprocessing
This study utilized cervical cancer screening public dataset provided by Intel and MobileODT that is hosted on the Kaggle portal [20]. The dataset comprised 3715 images in the training dataset and 452 images in the testing data sets. To create a validation dataset, we split the training dataset by 20%, resulting in 2973 images in the training set and 742 images in the validation set. The images’ resolution ranged from 1024 × 1024 to 3264 × 2448 pixels. However, due to hardware constraints, all images were resized to a uniform size of 256 × 256 and also normalized using Z-score normalization before feeding them into the CNN model.
Convolutional neural networks
Convolutional Neural Networks (CNNs) have become a prominent tool in medical imaging and have shown promising results in various applications, including, to mention but a few, cancer detection and screening, heart anomalies and Tuberculosis (TB) diagnosis [21-24]. In this research, we employed a deep- learning CNN to classify cervical cancerous and non-cancerous images, leveraging the power of this technology to improve the accuracy and speed of diagnosis.
A CNN is a deep neural algorithm inspired by the working operation of an animal visual cortex and it is primarily used for image and video processing that applies a set of learnable filters to the input to extract and detect specific features, which are then used to perform classification or regression tasks [25]. It consists of an input and output layer with multiple hidden layers stacked in the middle. These hidden layers are either convolutional, pooling or fully connected layers.
Convolutional layer: The convolutional layer is a fundamental building block of CNNs used for extracting various features from an input image using a convolutional filter, a small weight matrix applied to each part of the image to generate a corresponding feature map [26]. Each neuron in the layer processes data for its receptive field, allowing it to capture local features like edges, corners and textures. The convolutional layer reduces the number of free parameters in the network and enhances its generalization ability by sharing weights across neurons, preventing overfitting to training data. The equation shows the mathematical expression for the convolution operation.
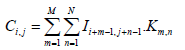
Where I is the input image, K is the convolutional filter, C is the output feature map and M and N are the dimensions of the filter. The value of Ci,j represents the result of applying the filter to the local receptive field of the input image centered at pixel (i,j). The dot product of the filter values Km,n and the corresponding input image values Ii+m-1,j+n-1 is computed for each element in the receptive field and the results are summed to produce the output value Ci,j. This operation is performed for all pixels in the input image, generating a complete output feature map that captures the local features of the image.
Pooling layer: The purpose of a pooling layer is to down sample the output of the previous convolutional layer, reducing its spatial dimensions while retaining important information. This process helps to decrease the computational complexity of the network and prevent overfitting [27]. The most common types of pooling layers are max pooling, which selects the maximum value in each pool and average pooling, which takes the average value. The pooling layer is typically followed by another convolutional layer, which continues to extract higher-level features from the down sampled output.
Fully connected layer: A fully connected layer, also known as a dense layer, is a type of neural network layer where each neuron in the layer is connected to every neuron in the previous layer. Each connection has a weight, which is learned during training and is used to compute a weighted sum of the inputs. This weighted sum is then passed through the neuron’s activation function to produce an output. The purpose of the fully connected layer is to learn complex relationships between the inputs and produce a final output, which can be used for either classification or regression [28].
Experimental setup
Development environment: The algorithm was developed in Python, utilizing the TensorFlow 2.11.0 and Keras 2.12.0 frameworks. We trained it on an MSI GL75 Leopard 10SFR laptop equipped with an 8GB NVIDIA RTX 2070 GDDR6 Graphical Processing Unit (GPU), using the Compute Unified Device Architecture (CUDA) 12.1 and CUDA Deep Neural Network (cuDNN) Software Development Kit (SDK) 8.7.0 platforms.
Architecture: In our study, we propose a CNN architecture comprising twenty-one (21) layers. This architecture includes seven convolutional layers with varying filter sizes and the same activation function (relu), six max-pooling layers for reducing the spatial dimensions of the feature maps, and a single fully connected layer with a dropout rate of 0.5. Table 1 displays the CNN architecture. The binary classification output layer employs a sigmoid activation function. We benchmarked this model on the VGG16 architecture and fine-tuned the parameters through numerous experimental tests until satisfactory results were obtained [29]. To prevent overfitting, we incorporated batch normalization and dropout in some of the hidden layers and we also utilized an L2 regularization of 0.0005 in all the hidden layers. The model had a total number of 1,889,152 parameters, 1,886,913 trainable parameters and 2,240 non-trainable parameters (Table 1).
| Layer no. | Layer type | Filter size | Stride | No. of filters | Padding | Fc units | Input | Output | Parameters |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Convolution 1 | 3 × 3 | 3 × 3 | 32 | valid | - | 256 × 256 × 3 | 85 × 85 × 32 | 896 |
| 2 | Max pooling 1 | 2 × 2 | - | - | - | - | 85 × 85 ×32 | 42 × 42 × 32 | 0 |
| 3 | Batch norm 1 | - | - | - | - | - | 42 × 42 × 32 | 42 × 42 × 32 | 128 |
| 4 | Convolution 2 | 3 × 3 | 1 × 1 | 64 | same | - | 42 × 42 × 32 | 42 × 42× 64 | 18496 |
| 5 | Max pooling 2 | 2 × 2 | 2 × 2 | - | - | - | 42 × 42 × 64 | 21 × 21× 64 | 0 |
| 6 | Batch norm 2 | - | - | - | - | - | 21 × 21 × 64 | 21 × 21 × 64 | 256 |
| 7 | Convolution 3 | 3 × 3 | 1 × 1 | 64 | same | - | 21 × 21 × 64 | 21 × 21 × 64 | 36928 |
| 8 | Max pooling 3 | 2 × 2 | 2 × 2 | - | - | - | 21 × 21 × 64 | 10 × 10 × 64 | 0 |
| 9 | Convolution 4 | 3 × 3 | 1 × 1 | 128 | same | - | 10 × 10 × 64 | 10 × 10 × 128 | 73856 |
| 10 | Max pooling 4 | 2 × 2 | 2 × 2 | - | - | - | 10 × 10 × 128 | 5 × 5 × 128 | 0 |
| 11 | Batch norm 3 | - | - | - | - | - | 5 × 5 × 128 | 5 × 5 × 128 | 512 |
| 12 | Convolution 5 | 3 × 3 | 1 × 1 | 128 | same | - | 5 × 5 × 128 | 2 × 2 × 128 | 147584 |
| 13 | Max pooling 5 | 2 × 2 | 2 × 2 | - | - | - | 2 × 2 × 128 | 2 × 2 × 128 | 0 |
| 14 | Batch norm 4 | - | - | - | 2 × 2 × 128 | 2 × 2 × 128 | 512 | ||
| 15 | Convolution 6 | 3 × 3 | 1 × 1 | 256 | same | - | 2 × 2 × 256 | 2 × 2 × 256 | 295168 |
| 16 | Max pooling 6 | 2 × 2 | 2 × 2 | - | - | - | 2 × 2 × 256 | 1 × 1 × 256 | 0 |
| 17 | Batch norm 5 | - | - | - | - | - | 1 × 1 × 256 | 1 × 1 × 256 | 1024 |
| 18 | Convolution 7 | 3 × 3 | 1 × 1 | 512 | same | - | 1 × 1 × 256 | 1 × 1 × 512 | 1180160 |
| 19 | Batch norm 6 | - | - | - | - | - | 1 × 1 × 512 | 1 × 1 × 512 | 2048 |
| 20 | Dense 1 | - | - | - | - | 256 | - | - | 131328 |
| 21 | Dropout (0.5) output | - | - | - | - | 1 | - | - | 257 |
Table 1: Description of the convolutional neural network architecture.
Model implementation: The proposed CNN architecture was used to train the model on a pre-processed dataset resized to 256 × 256 and padded. The training process was performed for multiple epochs and the outcomes for different epochs are present in Table 2. It took approximately 6 minutes and 34 seconds to complete one epoch of the training process. The overall run time for the entire training process depended on the number of epochs specified during training. During the training process, the Adam optimizer was used with backpropagation to minimize the loss function, which measures the difference between the predicted outputs and the actual labels of the training and validation data sets. A learning rate of 0.0001 and a momentum of 0.9 were used for the optimizer.
| Epochs | Accuracy | Precision | Recall | F1-score | Area Under Curve (AUC) |
|---|---|---|---|---|---|
| 20 | 82.74 | 80.91 | 85.90 | 83.33 | 88.10 |
| 25 | 91.37 | 88.80 | 94.69 | 91.65 | 96.00 |
| 50 | 87.00 | 85.00 | 90.00 | 88.00 | 92.00 |
| 90 | 89.50 | 87.50 | 92.00 | 89.54 | 94.10 |
Table 2: Performance metrics of the model for different epochs.
Evaluation metrics: The performance of the proposed cervical cancer screening algorithm was evaluated using standard classification metrics, including accuracy, precision, recall, F-score and area under the curve. These metrics are commonly used to assess the performance of binary classifiers like the one developed in this study.
• Accuracy: Accuracy is the most commonly used metric for evaluating the performance of a binary classifier. It is defined as the ratio of correct predictions to the total number of predictions made by the model:
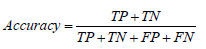
• Precision: Precision, also known as specificity or False Positive Rate (FPR), is a metric that measures the proportion of positive predictions made by the model that is actually correct:
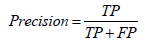
• Recall: Recall, also known as sensitivity or true positive rate, is a metric that measures the proportion of actual positive samples that the model correctly classifies:
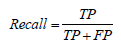
• F-score: The F-score, also known as the F1 score, is a metric that combines precision and recall into a single score:
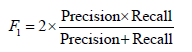
In the context of cervical cancer screening, we define True Positive (TP) as the number of correctly classified images that show signs of pre-cancerous cells, while True Negative (TN) represents the number of correctly classified images that are free of pre-cancerous cells. False Positive (FP) represents the number of images that are incorrectly classified as pre-cancerous when they are actually negative and False Negative (FN) represents the number of images that are incorrectly classified as negative when they are actually pre-cancerous.
• Area under the curve: The Area Under the Curve (AUC), takes into account both the sensitivity (True Positive Rate (TPR)) and specificity (False Positive Rate (FPR)) of the model, making it particularly useful for evaluating complex models like CNNs that involve large amounts of data and feature extraction. A good model should have a high TPR and a low FPR, indicating that it can accurately identify positive samples while minimizing the number of false positives. Mathematically, AUC is calculated by plotting the TPR against the FPR over different threshold values and measuring the area under the resulting curve, which ranges from 0.5 to 1.0, with higher scores indicating better performance. It is mathematically expressed as below.
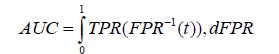
Where; t is the threshold value used to classify the samples, which is used to calculate TPR and FPR at different points on the ROC curve and dFPR is the infinitesimal change in the false positive rate used to compute the integral over the range of FPR values from 0 to 1.
In this study, we propose a CNN architecture for the task of screening cervical cancer by accurately classifying between normal and precancerous cervix. Our approach is especially relevant in countries like Uganda, where there are very few well-trained personnel to perform screening and diagnosis of cervical cancer. The shortage of skilled personnel can make it challenging to achieve high accuracy in screening and diagnosis. Our proposed approach offers a potential solution to this problem by providing an efficient and accurate method for cervical cancer screening, even in resource-constrained settings.
The objective of this study was to explore whether deep learning CNN models could effectively conduct cervical cancer screening tests and, if so, to determine the most appropriate CNN architecture and parameters for this purpose. The results indicate that satisfactory outcomes were achieved using the architecture presented in Table 1 and the training parameters are outlined.
We used a threshold of 0.5 to convert network output probabilities to binary values and the performance of the model was evaluated using various metrics such as accuracy, precision, recall, F1 score and AUC. The results are presented in Table 2 and the confusion matrix is shown in Figure 2. The model trained with 25 epochs achieved the best performance with an accuracy of 91.37%, precision of 88.80%, recall of 94.69%, an F1 score of 91.65%, and AUC of 96.0% as shown in Figure 3. These results suggest that our proposed CNN architecture is effective in accurately classifying the images (Table 2).
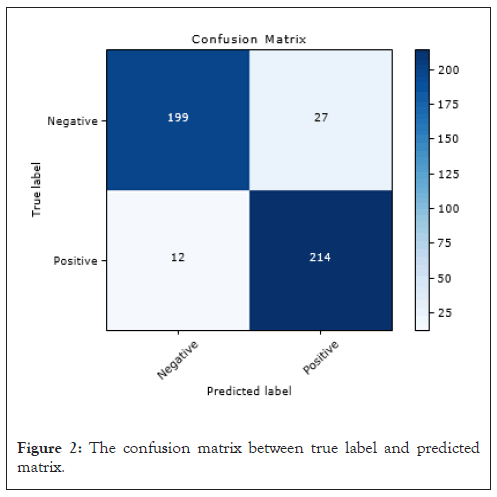
Figure 2: The confusion matrix between true label and predicted matrix.
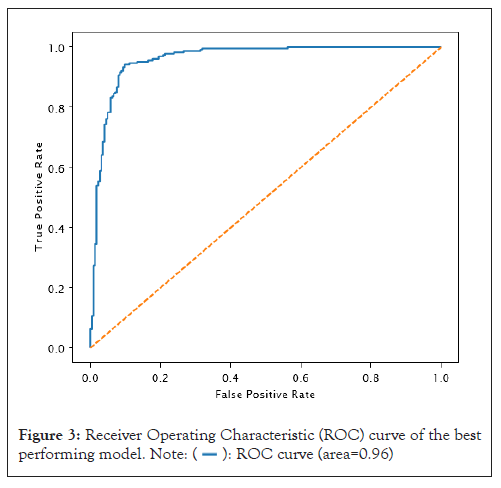
Figure 3: Receiver Operating Characteristic (ROC) curve of the best
performing model.  .
.
Figure 2 displays the confusion matrix resulting from applying our top performing model to the testing dataset, which consisted of 452 images. The algorithm accurately identified 413 images, correctly classifying 199 as normal and 214 as containing cancerous lesions. However, we observed that the algorithm misclassified 39 images. Of particular concern are the 12 false negatives, as they represent a significant risk. Failing to detect cancer when present can lead to more severe consequences, as cancer may progress to more advanced stages (Figure 2).
We generated a Receiver Operating Characteristic (ROC) curve to evaluate the performance of our model in distinguishing true positives from negatives, as illustrated in Figure 3. The ROC curve is critical as it measures the ability of the CNN model to classify both positive and negative samples correctly. We derived the curve by graphing the true positive rate against the false positive rate at different threshold values. Notably, the AUC of our model was 0.96, indicating an excellent ability to distinguish between true positives and negatives (Figure 3).
In this study, we have introduced an approach for cervical cancer screening that utilizes a 21-layer deep learning CNN architecture. Our approach effectively distinguishes between normal and precancerous cervix, providing a potential solution to the issue of limited skilled personnel available for cervical cancer screening and diagnosis in Uganda. With an accuracy of 91.37%, a precision of 88.80%, a recall of 94.69%, an F1 score of 91.65% and an AUC of 96.0%, the algorithm exhibits high sensitivity and specificity in identifying potential cervical cancer cases, concluded that CNN algorithm developed in this research could be adopted for clinical deployment.
The findings of this study highlight the potential of deep learning CNN algorithms in improving cervical cancer screening, and further research will focus on deploying the model into Uganda EMR for clinical applications and evaluation of the algorithm performance on locally collected data.
We are so grateful to the Intel and MobileODT cervical cancer screening project for providing us with the image dataset.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Mirugwe A, Ashaba C (2023) Cervical Cancer Screening: Artificial Intelligence Algorithm for Automatic Diagnostic Support. Trans Med. 13:309.
Received: 17-Nov-2023, Manuscript No. TMCR-23-28055; Editor assigned: 20-Nov-2023, Pre QC No. TMCR-23-28055 (PQ); Reviewed: 04-Dec-2023, QC No. TMCR-23-28055; Revised: 11-Dec-2023, Manuscript No. TMCR-23-28055 (R); Published: 18-Dec-2023 , DOI: 10.35248/2161-1025.23.13.309
Copyright: © 2023 Mirugwe A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.