Review Article - (2018) Volume 4, Issue 3
Biosensors: A Novel Approach to Detect Food-borne Pathogens.
*Corresponding Author: Pınar Sanlibaba, Department of Food Engineering, Faculty of Engineering, Ankara University, 50 Year Campus 06830 Golbasi, Ankara, Turkey Email:
Abstract
Foodborne pathogens affect human health negatively and are known to cause economic losses. Therefore, quick detection of foodborne pathogens and the implementation of measures to ensure their inactivation are of immense significance. Immunological, molecular, and cultural methods are frequently used in the detection of foodborne pathogens. High cost, prolonged analysis times, and the necessity of specialized personnel are some of the disadvantages of these methods. Biosensors are known as analytical devices. The use of biosensors is considered a new approach to quickly detect foodborne pathogens and their toxins. Biosensors, which are capable of converting biological, chemical, or biochemical signals into measurable electrical signals, are systems containing a biological detection material combined with a chemical or physical transducer. Different types of biosensor are being employed for detection of pathogenic bacteria. Biosensors are sensitive, fast, economical, reliable, and portable devices, and are used in many fields such as food safety, medicine, pharmacy, measurement of environmental pollution, and the military defense. Electrochemical and optical biosensors and piezoelectric immunosensors are among the most frequently used biosensors in the detection of foodborne pathogens. In this article, the principle components and requirements for an ideal biosensor, types, and their applications in food industry are summarized.
Keywords: Food safety; Microbial biosensors; Pathogens detection; Rapid measurement
Introduction
Foodborne illness is one of the significant public health problems worldwide. Therefore, microbiological safety of food has become an important concern for consumers, various industries, and regulatory agencies [1]. There are many different groups of microorganisms in food. Some of these microorganisms maintain their normal life functions in food and are used in food production, whereas others may cause food spoilage or foodborne diseases. The most important pathogens found in food are Salmonella spp., Campylobacter spp., some strains of Staphylococcus aureus, Listeria monocytogenes, Bacillus cereus, Bacillus anthracis (produces anthrax toxin), Clostridium spp., Escherichia coli O157:H7, Shigella spp., Yersinia enterocolitica, Vibrio cholera, Brucella spp., Aeromonas spp. and Coxiella burnetii [2]. These bacteria mostly produce toxins and other cell metabolites that cause deadly diseases [3]. The period of analysis, high cost, and necessity of expert personnel limit the use of existing detection methods. Therefore, researchers focus on developing methods that are user-friendly, easy, precise, portable, cheap, rapid, and provide simultaneous results in the detection of pathogens [4]. There are four major categories of methods for detecting foodborne pathogens: (i) culture-based conventional microbiological methods, (ii) polymerase chain reaction (PCR), (iii) enzyme-linked immunosorbent assay (ELISA), and (iv) microarray-based techniques. The conventional microbiological methods of detection are considered to be the “gold-standard” and are well known for their cost-effectiveness, sensitivity, ability to confirm cell viability, and ease of standardization. However, it takes two or three days for the detection and up to 7-10 days for confirmation. Although PCR detection of different foodborne pathogens has been proven to be an invaluable method; real-time PCR is the most commonly used technique for quantification of specific DNA fragments. PCR is a rapid and sensitive method. Sometimes, false-negative or false-positive results are obtained and further confirmation is needed. ELISA is accurate, precise, and also ideal for qualitative and quantitative detection of many types of proteins in a complex matrix. Its sensitivity is low and it takes about 3-4 h to complete. The most recent group is microarray-based techniques. These methods have some advantages such as being informative, highly repeatable and possess the potential to combine detection, effectively identify, and quantify an unlimited number of foodborne pathogens in a single experiment. However, expensive equipment for array scanning and data collection are needed in this method [5,6].
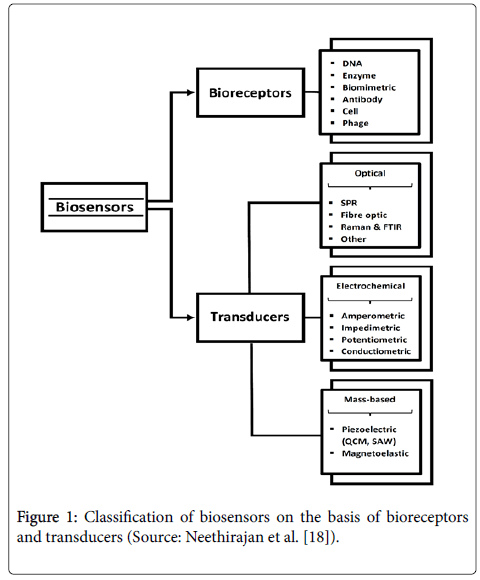
Rapid detection methods of foodborne pathogens can be categorized into nucleic acid-, antigen-antibody-based, biosensor-based, and bacteriophage-based methods. Biosensor-based methods have been increasingly gaining popularity owing to their characteristic feature of rapid detection of foodborne pathogens [1]. In addition to the rapid results, online biosensor technology offers the food industry a tool for internal process control to fulfil the high standard of quality control [7]. The application of biosensor technology offers promising solutions for portable, rapid, and sensitive detection of microorganisms in the food industry [4]. The history of biosensors dates back to as early as 1906. The first “true biosensor” was characterized by Leland C. Clark and Lyons in 1956 for oxygen detection. Leland C. Clark is known as the “father of biosensors” and his invention of the oxygen electrode bears his name “Clark electrode.” The first commercial biosensor was developed by Yellow Spring Instruments in 1975 [8]. In 1977, Rchenitz used the term “Bio selective sensor.” At a later stage, this term was abbreviated to “biosensor.” Biosensors mainly consist of two parts, viz., bioreceptors and transducers (Figure 1). The first part is a section where a specific biological event for recognition occurs. Bioreceptors are capable of binding to a specific substrate and can be grouped into five distinct classes, namely, antibody-antigen, enzymatic, nucleic acid, cellular, biomimetic, and bacteriophagic bioreceptors [7]. Some biological molecules such as antibodies, enzymes, proteins, nucleic acids, and viable biological systems such as cells, tissues, and microorganisms can be used as bioreceptors [9]. Enzymes, antibodies, and nucleic acids are the main classes of bioreceptors [10]. The second part is a transformer system that converts the biological reaction into a measurable signal [11]. This part plays a crucial role in the detection and identification process of a biosensor [12]. Biosensors, which are capable of converting biological, chemical, or biochemical signals into measurable electrical signals, are systems containing a biological detection material combined with a chemical or physical transducer. Various biological identification elements are involved in biosensors. The transducer is responsible for ensuring that the signal is transmitted from the output area of the bioreceptors to the electrical field [13]. Biosensors can also be classified based on the transduction methods. There are new types of transducers being developed to be used as a part of biosensors. However, optical, electrochemical, and mass-sensitive transduction methods are given importance as these are the most common methods [10].
Figure 1: Classification of biosensors on the basis of bioreceptors and transducers (Source: Neethirajan et al. [18]).
This developing technology of biosensors is being used in the detection of biological and chemical agents in the fields of food analysis, agricultural production, environmental pollution, medicine, pharmacy, mining, biotechnology, military defense, and country security [14]. We aimed to discuss and summarize various types of biosensors and their applications in detecting foodborne pathogens in this article.
Biosensors Used In The Food Industry For Detecting Pathogens
The food industry constantly seeks to improve production, feasibility, and quality to reduce production costs and time, and to conduct effective quality-control methods to satisfy the consumer [11]. Biosensors have been developed as important alternatives to traditional methods to ensure quality and safety in the food processing industry in a fast, precise, and easy manner. Biosensors developed for the food sector have been used in many applications such as quality control of food components and the detection of microbial and/or chemical ingredients for food safety [15].
Common food such as milk, cheese, meat, chicken, raw vegetables, and fruits are contaminated with pathogenic microorganisms. The traditional methods of detection need around 1-2 days to determine the pathogens. The use of biosensors is the best upcoming technology to combat this problem [3]. There are some advantages of using biosensors in the food industry. First, biosensors are frequently used in the determination of many substances such as glucose, monosaccharides, amino acids, organic acids, urea, and alcohol. Second, they are used to determine parameters such as aroma and freshness and to detect drugs and other such material in foods. Moreover, in environmental tracking, biosensors are used to determine pesticidal and antibiotic residues, toxins, and microorganisms and to measure biochemical oxygen demand (BOD) in the air, water, and soil samples [16]. Besides, various enzymes such as glucose oxidase, urease, and peroxidase have been widely used to amplify biological signals for improving the sensitivity of the biosensors in the detection of foodborne pathogens or other small biomolecules [17].
Various types of biosensors have been characterized. In general, biosensors can be divided into two groups, viz., direct and indirect biosensors [18]. Direct detection sensors are non-catalytic elements such as cell receptors or antibodies. Biological interactions are directly measured in real time in the direct detection sensors. Indirect detection sensors rely on a primary recognition reaction that binds the analyte to a substrate followed by a secondary recognition reaction that binds antibodies as the recognition element called as immunosensors. Although direct-detection biosensors are simpler and faster, they typically yield a higher limit of detection than indirect-detection systems [19].
An ideal biosensor should have a high selectivity for the target analyte (should not tend to bind with or have an affinity toward other reagents) and should be sensitive to the change in the amount of substance to be measured. At the same time, the biological material mobilized as the biosensor should be sensitive only to certain substances. An ideal biosensor should have high electrode stability. This depends on the physical strength of the biological material used. An ideal biosensor should always give the same results for the same sample concentrations in multiple measurements [20]. For a biosensor to work effectively, it is necessary to respond quickly in real-time tracking of the target analyte. However, the characteristics of an ideal biosensor are that it should be precise, repeatable, and linear. It should not give false-negative results, and the false-positive results should be minimal. Generally, ideal biosensors are automated systems and should require minimal operator intervention, have a simple design, and be inexpensive, easy to use, small, and portable [15]. Types of biosensors for the detection of foodborne pathogen are summarized in Table 1.
| Target microorganism | Food Sample | Biosensor | Detection Limit | References |
|---|---|---|---|---|
| Staphylococcus aureus | Buffer milk | Fluorescence resonance energy transfer based | 1.5 × 102 cells/mL | [21] |
| Raw milk | Colorimetric (gold nanaparticle based) | 101-106 CFU/mL | [22] | |
| Chicken | Colorimetric immunosensor | 10 CFU/ml | [23] | |
| Food, environmental and biological samples | Electrochemiluminescent | 3.1 × 102 CFU/mL | [24] | |
| Spiked milk | Electrochemical immunosensor | 13 CFU/mL | [25] | |
| Milk, cheese and meat | Amperometric immunosensor | 10 CFU/mL | [26] | |
| Pig skin | Potentiometric | 2.4 × 103-2.0 × 104 CFU/mL | [27] | |
| Pork | Surface-enhanced Raman spectroscopy (SERS) aptasensors | 102-107 CFU/mL | [28] | |
| Culture and milk | Piezoelectric | 4.1 × 101-4.1 × 105 CFU/mL | [29] | |
| Culture | Quartz Crystal Microbalance with dissipation tracking (QCM-D) | 104 CFU/mL | [30] | |
| Spinach leaves | Magnetoelastic | 1.0 × 101-1.0 × 108 CFU/25 mm2 surface of spinach | [31] | |
| Culture | Magnetoelastic immunosensor | 104-108 CFU/mL | [32] | |
| Fresh fish and water | Impedimetric aptosensor | 10-106 CFU/mL | [33] | |
| Culture | Immonosensor | 101 CFU/mL | [34] | |
| Salmonella typhimurium |
Tomato surface | Magnetoelastic | 5 × 101-5 × 108 CFU/mL | [35] |
| Pork | Surface-enhanced Raman spectroscopy (SERS) aptasensors | 102-107 CFU/mL | [28] | |
| Culture | Quartz Crystal Microbalance (QCM) based aptasensor | 103 CFU/mL | [36] | |
| Chicken-rinse water | Electrochemical immonosensor | 1.04 × 103 CFU/g | [37] | |
| Chicken breast | Microfluidic-based nano-biosensor | 103 CFU/mL | [38] | |
| Milk | Amperometric | 10 CFU/mL | [39] | |
| Apple juice | Aptosensors (label-free) | 102-108 CFU/mL | [40] | |
| Salmonella pullorum |
Eggs and chicken meat | Electrochemical immunosensor (sandwich) | 3.0 × 103 CFU/mL | [41] |
| Salmonella enteritidis | Milk | Surface plasmon resonance (SPR) | 1 × 102 CFU/mL | [42] |
| Salmonella ATCC 50761 | Physiological saline | Aptosensors (label-free) | 75 and 7.5 × 105 CFU/mL | [43] |
| Salmonella gallinarum |
Eggs and chicken meat | Electrochemical immunosensor (sandwich) | 3.0 × 103 CFU/mL | [41] |
| E.coli O157:H7 | Ground beef | Electrochemical immunosensor | 2.05 × 103 CFU/g | [37] |
| Yoghurt | Smartphone-based fluorescence | 1 CFU/mL | [44] | |
| Culture | Surface plasmon resonance (SPR) | 0.6 × 106 CFU/mL | [6] | |
| Egg | Smartphone-based fluorescence | 10 CFU/mL | [44] | |
| Culture | Aptasensor based | 105 CFU/mL | [45] | |
| Milk and water | Antibody-based immunosensor | 100-105 CFU/mL | [46] | |
| E.coli | Drinking water | Fluorescence based | Less than 10 cells | [47] |
| Listeria monocytogenes |
Culture | Surface plasmon resonance (SPR) | 0.7 × 107 CFU/mL | [6] |
| Milk | Piezoelectric | 102 CFU/mL | [48] | |
| Spiked milk | Colorimetric | 11.7 × 102 CFU/mL | [49] | |
| Campylobacter jejuni | Culture | Quartz Crystal Microbalance (QCM) immunosensor | 150 CFU/mL | [50] |
| Vibrio parahaemolyticus | Culture | Aptasensor (sandwich type) | 10 CFU/mL | [51] |
Table 1: Types of Biosensor for the detection of foodborne pathogens.
Immunosensors
Immunosensors are biosensors based on the interactions of specific antibodies with a specific antigen. Antigens detect the binding of antibodies to the antigen by immobilizing the reaction on the surface of a transducer that converts the surface change parameters into detectable electrical signals. Because the diffusion of the antigens to the immobilized antibodies is limited, in particular, it is difficult to detect small amounts of contaminants in real time by immunological reactions [52]. Immunological methods involve the use of monoclonal and polyclonal antibodies. ELISA and lateral flow immunoassay are among the immunological methods that are currently used for the detection of foodborne pathogens [53].
A sandwich immunoassay was worked out for two Salmonella species (S. gallinarum and S. pullorum) in eggs and chicken meat by Fei et al. [41]. Researchers reported that a linear response to the Salmonella species was obtained in the concentration range of 104-109 CFU/ mL, and the detection limit was 3.0 × 103 CFU/mL for both species. Immunosensors working with screen-printed interdigitated microelectrode (SP-IDME) transducers were studied by Xu et al. [37]. Their results showed that the immunosensor was capable of specifically detecting E. coli O157:H7 and S. typhimurium within the range of 102-106 CFU/mL in pure culture samples. E. coli O157:H7 in ground beef and S. typhimurium in chicken-rinsed water were also examined in their study. They found that the limits of detection for the two bacteria in the culture samples were 2.05 × 103 CFU/g and 1.04 × 103 CFU/mL, respectively. Silva et al. [54] used cadmium selective polymeric membrane microelectrode (Cd-ISE) as a transducer for the detection of S. typhimurium in milk. It was observed that the detection limit was 2 cells per 100 μL. The average total time per assay of 75 minutes for the detection of S. typhimurium in milk samples was reported in their research. The developed immunosensors was applied to detect stress and resuscitate bacteria by Bekir et al. [34]. A stable and reproducible immunosensors with a sensitivity of 15 kΩ/decade and a detection limit of 101 CFU/mL was obtained for S. aureus concentrations ranging from 101-107 CFU/mL in their study. They implied that a low deviation in the immunosensors response (± 10%) was observed when it was exposed to stressed and unstressed bacteria.
Enzyme-based biosensors: The first potentiometric enzyme biosensor was reported by Guilbault and Montalvo in 1969 for the measurement of glucose levels using immobilized glucose oxidase enzyme. Other enzyme electrodes were developed later on based on urease, glutamate dehydrogenase, and lactate dehydrogenase [3]. Enzyme as a bio receptor has many advantages on fluorescent and radiolabeled substances. The enzyme immunoassay reagents are stable, sensitive, and non-hazardous. The enzyme bio receptor is suitably bound to the transducer by immobilization. Enzyme immobilization is used as a basis for improving biosensor components with features such as storage stability, sensitivity, high selectivity, short response time, and high reproducibility. Pathogenic bacteria such as L. monocytogenes, E. coli, and C. jejuni can be detected by labeling the antibody with enzymes. The most commonly used enzymes are horseradish peroxidase (HRP) and beta-galactosidase [7]. Hesari et al. [47] developed a strategy for rapid detection of E. coli in drinking water. Their study was based on the use of the substrate 4-methylumbelliferyl-β-d-glucuronide (MUG), which is hydrolyzed rapidly by the action of E. coli β-d-glucuronidase (GUD) enzyme. Depending on the number of bacteria in the sample, they found that the detection time required for the biosensor response ranged between 20 and 120 minutes. GUD enzymatic response was also measured and determined to be less than 10 E. coli cells in a reaction vial in their study.
Optical biosensors
Fibre optics was the first commercially available optical biosensor in which pathogens or toxins are fluorescently labelled, which when bound to the surface of the biosensor gets excited by laser wave (635 nm) [3]. Optical biosensors are categorized by light mode used for the detection of an analyte or by light scattered by samples. Simple optical sensors use light emission and detect changes in light intensity or spectrum shift. This may occur due to an analyte or a specific antibody-antigen binding in the presence of a light source. Optical sensors can be categorized as absorbent sensors. UV-visible (including ultraviolet) light, infrared, evanescent area, surface plasmon resonance (SPR), including transmission in luminescence and photoemissions, use various optical mechanisms for detection. An optical biosensor is a compact analytical device that is integrated into an optical transducer system or includes a connected biological detection element. The biosensor principle is typically based on an enzyme system that transforms the analytes into products that can be oxidized or reduced to a catalytically working electrode and converted into products that can be stored at a certain potential. Optical biosensors are a powerful alternative to traditional analytical techniques with their high specificity and sensitivity as well as small size and cost-effectiveness [55]. A biosensor with a working range of 103-106 CFU/mL was used by Adak et al. [56] for the detection of S. aureus. A detection limit between 102-103 CFU/mL of S. aureus was observed in the culture.
Fluorescence resonance energy transfer-based biosensors: The fluorescence resonance energy transfer (FRET)-based biosensors is a device with radiation-free energy transfer from the donor to the receiver. The quantitative analysis of bio molecular dynamics and protein-protein interactions between protein and DNA, including conformational changes in proteins, can be performed by the FRET technology. The use of FRET-based biosensors has been extended to allow tracking of cellular dynamics in both heterogeneous cell populations and single-cell levels [57]. Fluorescence biosensors were used for the rapid detection of S. aureus in the buffer and spiked milk by He et al. [21] and their assay allowed the detection of microbes in a buffer and spiked milk at concentrations of S. aureus as low as 1.5 × 102 CFU/mL and 7.6 × 102 CFU/mL, respectively. Xue et al. [58] researched the proposed fluorescent biosensor using the double-layer channel with the immune magnetic nanoparticles (MNPs) for specific separation and efficient concentration of the target bacteria. This biosensor was demonstrated to be able to detect E. coli O157:H7 at a concentration as low as 14 CFU/mL within 2 h. The recovery of E. coli in the spiked milk samples ranged from 95.92% to 108.15%, indicating that it was capable of detecting E. coli in real samples. Zeinhom et al. [44] used a portable smartphone-based fluorescence device for E. coli O157:H7 detection in yoghurt and eggs. They found that the detection limits were 1 CFU/mL and 10 CFU/mL in yoghurt and eggs, respectively. Recovery percentages of spiked yogurt and egg samples with 103, 104, and 105 CFU/mL E. coli O157:H7 were found to be 106.98% and 96.52%, 102.65% and 107.37%, 105.64%, and 93.84% in yogurt and egg samples, respectively, using their device. They reported that the entire process could be completed within 2 h.
Surface plasmon resonance biosensors: Surface plasmon resonance (SPR) occurs when light is reflected on the inner surface of a material with varied refractive indices. Between two layers, a thin layer of a good conductor, such as gold or silver, with a specific energy to raise the surface plasmon is placed. SPR is a powerful tool that can measure the binding kinetics of two molecules without any fluorescent label [7,11]. SPR eliminates matrix turbidity by measuring the refractive index on the reverse side of the metal film in which the biological selective element is immobilized. SPR biosensors are used for the detection of foodborne pathogens [59]. It can also be used for the installation of immunosensors applied in the detection of food pathogens in various foods or food dilutions [60]. SPR was used for the detection of E. coli O157:H7, S. enteritidis, and L. monocytogenes by Zhang et al. [6]. The lower detection limits for E. coli O157:H7, S. enteritidis, and L. monocytogenes were determined to be 0.6 × 106, 1.8 × 106 and 0.7 × 107 CFU/mL, respectively, in the presence of nontarget pathogens at concentrations of 105-108 CFU/mL. Eser et al. [42] used the SPR technique for the detection of S. enteritidis in milk. The detection limit of the pathogens was found to be 1 × 102 CFU/mL in their study.
Colorimetric biosensors: The colorimetric method, which is an attractive optical method, allows rapid identification of the pathogens in the sample by colour change. Response signals can be seen and resolved with the naked eye without requiring any analytical tool [20]. A gold nanoparticle-based colorimetric aptasensor for S. aureus in raw milk was developed by Yuan et al. [22]. The concentration of S. aureus over the range from 101-106 CFU/mL was determined. The colorimetric sensor was tested with serial broth dilutions of Listeria bacteria by Alhogail et al. [49]. The lowest detection limit of the developed sensor for Listeria was found to be 2.17 × 102 CFU/mL within 30 s. The detection limit of the sensor in the spiked milk was 11.7 × 102 CFU/mL and in the spiked meat was 13.8 × 101 CFU/g detected within 15 minutes and without pre-enrichment steps. A colorimetric biosensor was used for the determination of S. aureus by Suaifan et al. [61]. Their experimental results showed detection limits as low as 7, 40, and 100 CFU/mL for S. aureus in pure broth culture, and that inoculated in food produces and environmental samples, respectively.
Electrochemical biosensors
Electrochemical detection methods are advanced transduction-based systems used for the identification and measurement of foodborne pathogens. Electrochemical biosensors measure an electrochemical response. They convert the occurring electrical signal directly into an electronic field and allow the development of compact system designs with simple instrumentation. They have some advantages over other analytical transduction systems. These are (i) comparable instrumental sensitivity, (ii) possibility to operate in turbid media, and (iii) possibility of miniaturization, which allows even small volumes to be analysed [3]. Electrochemical biosensors can be classified as amperometric, potentiometric, impedimetric, and conductometric biosensors [52,59]. Electrochemical biosensors are commonly used for detecting microorganisms in food [3]. A facile label-free electrochemiluminescent (ECL) biosensor was developed for the detection of S. aureus by Yue et al. [24]. The ECL intensity decreased linearly with S. aureus concentrations in the range of 1.0 × 103-1.0 × 109 CFU/mL, with a detection limit of 3.1 × 102 CFU/mL in that study. The author reported that the whole assay could be accomplished within 70 minutes when a ready-to-use biosensor was applied. The recovery test for food, environmental, and biological samples showed recoveries between 75.0% and 116.7%. An electrochemical immunosensor for label-free detection of S. aureus was studied by Bhardwaj et al. [25]. The authors implied that the biosensor with a rapid detection time (30 minutes) and a limit of detection of 13 CFU/mL in spiked milk samples can be used for rapid detection of pathogens in actual food samples with high sensitivity and specificity.
Amperometric biosensors: Amperometric transduction is a universal electrochemical detection method that is well used for pathogen detection. Amperometric biosensors are used to examine electrochemical reactions while measuring the current change in a constant potential. The analyte concentration in a solution is proportional to the response of the biosensors. Amperometric biosensors have the advantages of being extremely sensitive, fast, and inexpensive and are used to identify important foodborne pathogens such as E. coli O157:H7, Salmonella, L. monocytogenes, and C. jejuni [57]. Amperometric biosensors can work in two or three electrode configurations. These biosensors are used as immunosensors or genosensors for the detection of foodborne pathogens [3]. An amperometric immunosensor for the detection of S. aureus in food samples was devised by Majumdar et al [26]. The changes were quantified by the increase in amperometric response. The response of the sensors to increasing concentrations (101-108 CFU/mL) of a pure culture of S. aureus NCIM 2602 as well as S. aureus inoculated food samples (milk, cheese, and meat) was studied and a similar response pattern was observed for all the samples. The detection limit was decreased down to 10 CFU/mL in their study. An amperometric biosensor for S. typhimurium detection in milk was used by Alexandre et al. [39]. The biosensor device showed a qualitative behavior with a very low limit of detection of 101 CFU/mL and a detection time of 125 minutes.
Potentiometric biosensors: Leland Clark in 1962 discovered the first potentiometric biosensor to detect urea in 1969 [8]. Potentiometric biosensors are based on the measurement of oxidation and reduction potential of an electrochemical reaction. Thus, a pH-meter consists of an immobilized enzyme membrane surrounding the probe, where the hydrogen ions are produced or absorbed by the catalysed reaction. Potentiometric biosensors include the use of ion-selective electrodes to convert the biological reaction into an electrical signal. Potentiometric biosensors measure potential differences in conditions of below zero. Antibody-antigen binding causes a small change in charge of proteins that can be determined potentiometrically, and the method is not very sensitive because of the very small load. Recent potentiometric devices are based on field-effect transistor (FET) devices [11]. The potentiometric biosensor is used for the E. coli assay allowing a detection limit of as low as 10 cells/mL. The poor selectivity in some food samples is a major disadvantage associated with this biosensor [3]. E. coli, S. aureus, and S. epidermidis were determined in pig skin by potentiometric biosensors based on carbon nanotubes and aptamers, with a working range 2.4 × 103-2.0 × 104 CFU/mL by Zelada-Guillen et al. [27].
Impedimetric biosensors: Impedimetric biosensors are powerful systems used for the detection of electrochemical systems [3]. The impedance is defined as the resistance in the electric current against an alternating current in an electrical circuit. In principle, the impedance biosensors are based on changes in the conductivity of the environment through microbial metabolism of electrically charged ionic compounds and inert substrates of acidic products such as amino acid, lactic acid, and acetic acid. The connection of impedance with biological recognition technology to detect pathogens has led to the development of impedance biosensors, which have been widely used in recent years [19,62]. Sheikhzadeh et al. [40] have used aptosensors (label-free) that have a working range 102-108 CFU/mL for the rapid detection of S. typhi in apple juice. Similarly, Jia et al. [43] detected Salmonella (ATCC 50761) in physiological saline with glassy carbon electrode (GCE) transducer and aptosensor (label-free) operating between 75 and 7.5 × 105 CFU/mL. In another study, an impedimetric aptasensor operating in the range 10-106 CFU/mL was used to determine the presence of S. aureus (ATCC 29213) in a culture. Zhang et al. [28] used Surface-enhanced Raman spectroscopy (SERS) aptasensors in the 102-107 CFU/mL range in the determination of S. aureus and S. typhimurium in pork.
Mass-sensitive biosensors
Mass-sensitive biosensors are based on the transduction method, which contains minor changes in the biosensor mass. They are also known as piezoelectric biosensors because they are often used as piezoelectric crystals that can precisely determine small changes in the mass. They are less used than optical and electrochemical biosensors. The two main types of mass sensitive biosensors are the surface acoustic wave and the quartz crystal microbalance devices, also known as bulk wave devices [63,64].
Piezoelectric biosensors: The piezoelectric biosensors based on the principle of detecting bacteria directly without labelling are very interesting sensors. In general, the surface of the piezoelectric sensor is coated with a selective binding agent (e.g. antibodies) in which the bacteria-containing solution is placed. Bacteria bind to antibodies reducing the oscillation frequency as the crystal mass increases. Piezoelectric quartz crystal microbalance (QCM) is the main type of piezoelectric biosensor used in pathogen detection. QCM biosensors have advantages such as real-time monitoring, ease of use, unlabelled detection, and being biocompatible electrodes for ligand immobilization (such as Au) [55,57]. QCM biosensors are used in food, biochemistry, environment, and clinical fields and are similar to SPR biosensors in terms of selectivity and sensitivity, but need to be improved in terms of repeatability and stability [65]. A study conducted by Lian et al. [29] for the pathogen (S. aureus) detection in culture and milk, was carried out using piezoelectric biosensor and ranged between 4.1 × 101 and 4.1 × 105 CFU/mL. In another study conducted by Sharma and Mutharasan [48] using a piezoelectric biosensor, the number of L. monocytogenes in milk was found to be 102 CFU/mL. S. aureus was detected in a culture using Quartz Crystal Microbalance with dissipation tracking (QCM-D) by Guntupalli et al. [30] and the presence of S. aureus by using phage (Phage 12600) was found to be 104 CFU/mL. Wang et al. [66] also studied a QCM-based aptasensor that was developed to detect S. typhimurium. This aptasensor was able to detect 103 CFU/mL of S. typhimurium within 1 h.
Magnetoelastic biosensors: Magnetoelastic sensors are made from amorphous ferromagnetic alloys. Magnetoelastic sensors are characterized by remote sensing as the signal transmission is carried out at a distance from the coil. When stimulated by a magnetic field, which changes regularly, the materials exhibit a magnetoelastic resonance that can be determined by a noncontact signal collector coil. When a target is in contact with the pathogen alloy sensor surface, the added mass causes a change in the resonance frequency and can be detected remotely by the signal collector coil. Therefore, magnetoelastic sensors are wireless devices that can become very useful tools for remote monitoring. Magnetoelastic biosensors are the first example of wireless biosensors in biosensor platforms [55]. Byeon et al. [31] detected S. aureus in spinach leaves with magnetoelastic biosensor operating in the range 1.0 × 101-1.0 × 108 CFU/25 mm2 surface of spinach. Similarly, Menti et al. [32] used a magnetoelastic immunosensor in the range 104-108 CFU/mL to detect S. aureus in a culture. In a study, S. typhimurium was detected on the tomato surface using a magnetoelastic biosensor with a working range of 5 × 101-5 × 108 CFU/mL [35].
Conclusion
A great number of cases have been reported in recent years regarding foodborne pathogens, which may cause serious health problems or even death. Therefore, it is important to quickly detect such pathogens. Accordingly, several rapid analytical methods have been developed. One of these methods is the use of biosensors. The use of biosensors in the detection of foodborne pathogens is one of the promising methods in terms of their short analysis times, low costs, precision, and reliability. With the advancing technology, it is possible to develop more sensitive, faster, portable, comparable sensitive and economical biosensors. Therefore, further research is needed to develop biosensors that can detect foodborne pathogens and their toxins in a better way. Continuing research will reveal the best procedures and full applicability of whole-cell bacterial biosensors.
References
- Bavisetty SCB, Vu HTK, Benjakul S, Vongkamjan K (2018) Rapid pathogen detection tools in seafood safety. Curr Opin Food Sci 20: 92-99.
- Saglam D, Seker E (2016) Gida kaynakli bakteriyel patojenler. Kocatepe Vet J 9: 105-113.
- Majumdar T, Raychaudhuri U, Chakraborty R (2015) Detection of food borne pathogens. Int J Adv Biol Res 5: 96-107.
- Yamada K, Choi W, Lee I, Cho BK, Jun S (2016) Rapid detection of multiple foodborne pathogens using a nanoparticle-functionalized multi-junction biosensor. Biosens Bioelectron 77: 137-143.
- Bai S, Zhao J, Zhang Y, Huang W, Xu S, et al. (2010) Rapid and reliable detection of 11 food-borne pathogens using thin-film biosensor chips. Appl Microbiol Biotechnol 86: 983-990
- Zhang X, Kitaoka H, Tsuji S, Tamai M, Kobayashi H, et al. (2014) Development of a simultaneous detection method for foodborne pathogens using surface plasmon resonance biosensors. Food Sci Technol Res 20: 317-325.
- Sharma H, Agarwal M, Goswami M, Sharma A, Roy SK, et al. (2013) Biosensors: tool for food borne pathogen detection. Vet World 6: 968-973.
- Bhalla N, Jolly P, Formisano N, Estrela P (2016) Introductrion to bisensors. Essay Biochem 60: 1-8.
- Su L, Jia W, Hou C, Lei Y (2011) Microbial biosensors: A review. Biosens Bioelectron 26: 1788-1799.
- Velusamy V, Arshak K, Korostynska O, Oliwa K, Adley C (2010) An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnol Adv 28: 232-254.
- Meshram BD, Agrawal AK, Adil S, Ranvir S, Sande KK (2018) Biosensor and its application in food and dairy industry: A review. Int J Curr Microbiol App Sci 7: 3305-3324.
- Singh R, Mukherjee MD, Sumana G, Gupta RK, Sood S, et al. (2014) Biosensors for pathogen detection: A smart approach toward clinical diagnosis. Sens Actuators B197: 385-404.
- Özoglu Ö, Ünal MA, Altuntas EG (2017) Biyosensörler: gida ve saglik alaninda laktat biyosensörleri. Turk J Life Sci 2:180-193.
- Maas MB, Perold WJ, Dicks LMT (2017) Biosensors for the detection of Escherichia coli. Water SA 43: 4.
- Thakur MS, Ragavan KV (2013) Biosensors in food processing. J Food Sci Technol 50: 625-641.
- Gunes G (2015) Use of amino acid sensitive biosensors as detectors in liquid cramotography and determination of amino acids.
- Huang F, Zhang H, Wang L, Lai W, Lin J (2018) A sensitive biosensor using double-layer capillary based immunomagnetic separation and invertase-nanocluster based signal amplification for rapid detection of foodborne pathogen. Biosens Bioelectron 100: 583-590.
- Neethirajan S, Ragavan V, Weng X, Chand R (2018) Biosensors for sustainable food engineering: challenges and perspectives. Biosensors 8: E23.
- Naik MK, Srinivas D, Sasi B, Jakeer Basha SK (2017) Biosensors in food processing-A review. Int J Pure App Biosci 5: 1219-1227.
- Rubab M, Shahbaz HM, Olaimat AN, Oh DH (2018) Biosensors for rapid and sensitive detection of Staphylococcus aureus in food. Biosens Bioelectron 105: 49-57.
- He X, Li Y, He D, Wang K, Shangguan J, et al. (2014) Aptamer-fluorescent silica nanoparticles bioconjugates based dual-color flow cytometry for specific detection of Staphylococcus aureus. J Biomed Nanotechnol 10: 1359-1368.
- Yuan J, Wu S, Duan N, Ma X, Xia Y, et al. (2014) A sensitive gold nanoparticle-based colorimetric aptasensor for Staphylococcus aureus. Talanta 127: 163-168.
- Alamer S, Chinnappan R, Zourob M (2017) Development of rapid immuno-based nanosensors for the detection of pathogenic bacteria in poultry processing plants. Procedia Technol 27: 23-26.
- Yue H, Zhou Y, Wang P, Wang X, Wang Z, et al. (2016) A facile label-free electrochemiluminescent biosensor for specific detection of Staphylococcus aureus utilizing the binding between immunoglobulin G and protein A. Talanta 153: 401-406.
- Bhardwaj J, Devarakonda S, Kumar S, Jang J (2017) Development of a paper-based electrochemical immunosensor using an antibody-single walled carbon nanotubes bio-conjugate modified electrode for label-free detection of foodborne pathogens. Sens Actuators B Chem 253: 115-123.
- Majumdar T, Chakraborty R, Raychaudhuri U (2013) Development of PEI-GA modified antibody based sensor for the detection of S. aureus in food samples. Food Biosci 4: 38-45.
- Zelada-Guillén GA, Sebastián-Avila JL, Blondeau P, Riu J, Rius FX (2012) Label-free detection of Staphylococcus aureus in skin using real-time potentiometric biosensors based on carbon nanotubes and aptamers. Biosens Bioelectron 31: 226-232.
- Zhang H, Ma X, Liu Y, Duan N, Wu S, et al. (2015) Gold nanoparticles enhanced SERS aptasensor for the simultaneous detection of Salmonella typhimurium and Staphylococcus aureus. Biosens Bioelectron 74: 872-877.
- Lian Y, He F, Wang H, Tong F (2015) A new aptamer/graphene interdigitated gold electrode piezoelectric sensor for rapid and specific detection of Staphylococcus aureus. Biosens Bioelectron 65: 314-319.
- Guntupalli R, Sorokulova I, Olsen E, Globa L, Pustovyy O, et al. (2013) Biosensor for detection of antibiotic resistant Staphylococcus bacteria. J Vis Exp 75: e50474.
- Byeon HM, Vodyanoy VJ, Oh JH, Kwon JH, Park MK (2015) Lytic phage-based magnetoelastic biosensors for on-site detection of methicillin-resistant Staphylococcus aureus on spinach leaves. J Electrochem Soc 162: B230-B235.
- Menti C, Beltrami M, Pozza MD, Martins ST, Henriques JAP, et al. (2017) Influence of antibody immobilization strategies on the analytical performance of a magneto-elastic immunosensor for Staphylococcus aureus detection. Mater Sci Eng C Mater Biol Appl 76: 1232-1239.
- Jia F, Duan N, Wu S, Ma X, Xia Y, et al. (2014) Impedimetric aptasensor for Staphylococcus aureus based on nanocomposite prepared from reduced graphene oxide and gold nanoparticles. Microchim Acta 181: 967-974.
- Bekir K, Barhoumi H, Braiek M, Chrouda A, Zine N, et al. (2015) Electrochemical impedance immunosensor for rapid detection of stressed pathogenic Staphylococcus aureus bacteria. Environ Sci Pollut Res Int 22: 15796-15803.
- Li S, Li Y, Chen H, Horikawa S, Shen W, et al. (2010) Direct detection of Salmonella typhimurium on fresh produce using phage-based magnetoelastic biosensors. Biosens Bioelectron 26: 1313-1319.
- Wang L, Wang R, Chen F, Jiang T, Wang H, et al. (2017) QCM-based aptamer selection and detection of Salmonella typhimurium. Food Chem 221: 776-782.
- Xu M, Wang R, Li Y (2016) Rapid detection of Escherichia coli O157:H7 and Salmonella Typhimurium in foods using an electrochemical immunosensor based on screen-printed interdigitated microelectrode and immunomagnetic separation. Talanta 148: 200-208.
- Kim G, Moon JH, Moh CY, Lim JG (2015) A microfluidic nano-biosensor for the detection of pathogenic Salmonella. Biosens Bioelectron 67: 243-247.
- Alexandre DL, Melo AMA, Furtado RF, Borges MF, Figueiredo EAT, et al. (2018) A Rapid and Specific Biosensor for Salmonella Typhimurium Detection in Milk. Food Bioprocess Technol 11: 748-756.
- Sheikhzadeh E, Chamsaz M, Turner APF, Jager EWH, Beni V (2016) Label-free impedimetric biosensor for Salmonella Typhimurium detection based on poly [pyrrole-co–3-carboxyl-pyrrole] copolymer supported aptamer. Biosens Bioelectron 80: 194-200.
- Fei JF, Dou WC, Zhao GY (2015) A sandwich electrochemical immunosensor for Salmonella pullorum and Salmonella gallinarum based on a screen-printed carbon electrode modified with an ionic liquid and electrodeposited gold nanoparticles. Microchim Acta 182: 2267-2275.
- Eser E, Ekiz OÖ, Çelik H, Sülek S, Dana A, et al. (2015) Rapid detection of foodborne pathogens by surface plasmon resonance biosensors. Int J Biosci Biochem Bioinforma 5: 329-325.
- Jia F, Duan N, Wu SJ, Dai RT, Wang ZP, et al. (2015) Impedimetric Salmonella aptasensor using a glassy carbon electrode modified with an electrodeposited composite consisting of reduced graphene oxide and carbon nanotubes. Microchim Acta 183: 337-344.
- Zeinhom MMA, Wang Y, Song Y, Zhu MJ, Lin Y, et al. (2018) A portable smart-phone device for rapid and sensitive detection of E. coli O157: H7 in yoghurt and egg. Biosens Bioelectron 99: 479-485.
- Wu WH, Li M, Wang Y, Ouyang HX, Wang L, et al. (2012) Aptasensors for rapid detection of Escherichia coli O157:H7 and Salmonella typhimurium. Nanoscale Res Lett 7: 658.
- Alocilja EC, Jain P, Pryg K (2016) Immunosensor for rapid extraction/detection of enteric pathogens. Technol 4: 194-200.
- Hesari N, Alum A, Elzein M, Abbaszadegan M (2016) A biosensor platform for rapid detection of E. coli in drinking water. Enzyme Microb Technol 83: 22-28.
- Sharma H, Mutharasan R (2013) Rapid and sensitive immunodetection of Listeria monocytogenes in milk using a novel piezoelectric cantilever sensor. Biosens Bioelectron 45: 158-162.
- Alhogail S, Suaifan GARY, Zourob M (2016) Rapid colorimetric sensing platform for the detection of Listeria monocytogenes foodborne pathogen. Biosens Bioelectron 86: 1061-1066.
- Masdor NA, Altintas Z, Tothill IE (2016) Sensitive detection of Campylobacter jejuni using nanoparticles enhanced QCM sensor. Biosens Bioelectron 78: 328-336.
- Wu W, Zhou M, He H, Liu C, Li P, et al. (2018) A sensitive aptasensor for the detection of Vibrio parahaemolyticus. Sensors and Actuators B: Chemical 272: 550-558.
- Zhao X, Lin C, Wang J, Oh DH (2014) Advances in rapid detection methods for foodborne pathogens. J Microbiol Biotechnol 24: 297-312.
- Law JW, Ab Mutalib NS, Chan KG, Lee LH (2015) Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front Microbiol 5: 770.
- Silva NFD, Magalhaes JMCS, Oliva-Teles MT, Delerue-Matos C (2015) A potentiometric magnetic immunoassay for rapid detection of Salmonella typhimurium. Anal Methods 7: 4008-4011.
- Wang Y, Salazar JK (2016) Culture-independent rapid detection methods for bacterial pathogens and toxins in food matrices. Compr Rev Food Sci Food Saf 15: 183-205.
- Adak AK, Boley JW, Lyvers DP, Chiu GT, Low PS, et al. (2013) Label-free detection of Staphylococcus aureus captured on immutable ligand arrays. ACS Appl Mater Interfaces 5: 6404-6411.
- Soni DK, Ahmad R, Dubey SK (2018) Biosensor for the detection of Listeria monocytogenes: emerging trends. Crit Rev Microbiol 44: 590-608.
- Xue L, Zheng L, Zhang H, Jin X, Lin J (2018) An ultrasensitive fluorescent biosensor using high gradient magnetic separation and quantum dots for fast detection of foodborne pathogenic bacteria. Sens Actuators B Chem 265: 318-325.
- Mortari A, Lorenzelli L (2014) Recent sensing technologies for pathogen detection in milk: A review. Biosens Bioelectron 60: 8-21.
- Poltronieri P, Mezzolla V, Primiceri E, Maruccio G (2014) Biosensors for the detection of food pathogens. Foods 3: 511-526
- Suaifan GA, Alhogail S, Zourob M (2017) Rapid and low-cost biosensor for the detection of Staphylococcus aureus. Biosens Bioelectron 90: 230-237.
- Fegade U, Sharma H, Bondhopadhyay B, Basu A, Attarde S, et al. (2014) Turn-on fluorescent dipodal chemosensor for nano-molar detection of Zn (2+): application in living cells imaging. Talanta 125: 418-424.
- Yang X, Kirsch J, Simonian A (2013) Campylobacter spp. detection in the 21st century: A review of the recent achievements in biosensor development. J Microbiol Methods 95: 48-56.
- Yasmin J, Ahmed MR, Cho BK (2016) Biosensors and their applications in food safety: A review. J Biosystems Eng 41: 240-254.
- Syam R, Justin Davis K, Pratheesh MD, Anoopraj R, Bunglavan SJ (2012) Biosensors: a novel approach for pathogen detection. Vet Scan 7: 14-18.
- Wang W, Mai Z, Chen Y, Wang J, Li L, et al. (2017) A label-free fiber optic SPR biosensor for specific detection of C-reactive protein. Scientific Reports 7: 16904
Copyright: © 2018 Sentürk E, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.