
Journal of Clinical Trials
Open Access
ISSN: 2167-0870

ISSN: 2167-0870
Research Article - (2021)Volume 11, Issue 6
Background: LINC00634 is highly expressed in esophageal cancer, and its depletion can suppress the viability and induce the apoptosis of esophageal cancer cells. However, there is a lack of studies that examine the relationship between LINC00634 expression and the clinic pathological features, survival outcomes, prognostic factors and tumor immune cell infiltration of Colorectal Carcinoma (CRC) patients.
Objective: We aim at investigating the role of LINC00634 in colorectal carcinoma.
Methods: We obtained data from the TCGA (The Cancer Genome Atlas) public database, GTEx (Genotype-Tissue Expression) database and clinical samples. Wilcoxon rank-sum test, Kruskal-Wallis test and logistic regression analysis were employed to assess the relationship between LINC00634 expression and the clinicopathological characteristics of CRC patients. Receiver Operating Characteristic (ROC) curve was constructed to evaluate the ability of LINC00634 for distinguishing between CRC patients and normal subjects based on the Area Under the Curve (AUC) score. Univariate and multivariate analyses were conducted to evaluate the association between prognostic factors and survival outcomes. Kaplan-Meier curves and Cox regression analysis were employed to determine the contribution of LINC00634 expression to the prognosis of colorectal carcinoma patients. Immune infiltration analysis and Gene Set Enrichment Analysis (GSEA) were conducted to identify the significantly involved functions of LINC00634. Finally, a nomogram was constructed for internal verification based on the Cox regression data.
Results: The expression of LINC00634 was upregulated in CRC patients, and markedly associated with N stage, residual tumor, pathological stage, and Overall Survival (OS) event. ROC curve showed that LINC00634 had strong diagnostic and prognostic abilities (AUC=0.74). The high expression of LINC00634 could predict poor disease specific survival (DSS; P=0.008) and poor overroll survival (OS;P<0.01). The expression of LINC00634 was independently associated with OS in CRC patients (P=0.019). GSEA and immune infiltration analysis demonstrated that LINC00634 expression was involved in gene transcription, epigenetic regulation and the functions of certain types of immune infiltrating cells. The c-index of the nomogram was 0.772 (95%CI: 0.744-0.799).
Conclusion: Our study reveals that LINC00634 can serve as a potential prognostic biomarker for CRC patients.
Noninvasive prenatal testing; Copy number variants; Sex chromosome abnormalities; Advanced maternal age
Colorectal Carcinoma (CRC) is a frequent malignant tumor in the clinic. The global cancer statistical analysis in 2020 shows that CRC ranks the third among cancers in the world, and the mortality rate ranks second among cancer-related deaths in both men and women [1]. Increasing incidence of CRC is mainly attributed to the changes in lifestyle and eating habits. Specifically,a high consumption of animal-derived food and a sedentary lifestyle can lead to a decrease in physical activity and an increase in body weight, which is independently associated with the risk of CRC [2]. Other risk factors include heavy drinking, smoking, and consumption of red or processed meats [3]. Despite the advancements of diagnosis technology and treatment, predicting the prognosis of CRC patients is still a major challenge. Therefore, finding new molecular markers associated with CRC is of utmost urgency for the improving diagnosis and prognosis of this disease.
Long intergenic non-coding RNAs (LINCRNAs) are non-coding RNAs with a transcript length of longer than 200 nt, which are not capable of encoding proteins. However, LINCRNAs can control gene expression at the epigenetic level, transcription level, and post-transcriptional level [4]. In recent years, LINCRNAs have been widely studied as potential key factors for cancer cell regulation. LINCRNAs are abnormally expressed in almost all cancers, and they play pivotal roles in promoting and maintaining the occurrence and development of tumors. This suggests the clinical potential of LINCRNAs as biomarkers and therapeutic targets [5,6]. Several studies have found that LINCRNAs can interact with RNA, proteins and lipids, and act as key signal transduction mediators in cancer-related signal transduction pathway, which eventually affect the angiogenesis, proliferation, migration and invasion of tumor cells [7-9]. A recent study has reported that LINC00634 is highly expressed in esophageal cancer and activates BCL2L1 to regulate cell viability and cell apoptosis by sponging miR-342-3p in esophageal cancer, thus promoting the malignant progression of esophageal cancer cells [10]. However, there are only few reports about the relationship between LINC00634 and colorectal carcinoma.
In this research, the expression data were retrieved from the Cancer Genome Atlas (TCGA) public database, GTEx database and clinical samples to compare the differential expression levels of LINC00634 between tumor tissues and normal colorectal tissues and to examine the association between LINC00634 expression and the clinicopathological features of CRC patients. Subsequently, the potential of LINC00634 for predicting the prognosis of CRC patients was evaluated. Moreover, bioinformatics analysis was conducted on LINC00634 upregulation and downregulation groups to reveal the underlying biological functions. Furthermore, we determined the association between LINC00634 expression and immune cell infiltration, and elucidated the underlying mechanism of LINC00634 in regulating the occurrence and development of CRC.
Ethics statement
The current study was approved by the Ethics Committee of Guangzhou First People’s Hospital and performed in strict accordance with the Declaration of Helsinki. All participants (including living and dead patients) or their legal guardian signed informed consent prior to enrollment. Extensive efforts were made to ensure minimal usage of animals as well as their suffering.
Data collection and bioinformatics analysis
The clinical data of TCGA CRC tissues (n=647) and matched normal colorectal tissues (n=51) were obtained from the TCGA database. To detect LINC00634 expression, the data for tumor tissues (n=383) were obtained from the TCGA database, while those for normal colorectal tissues (n=359) were combined with normal tissues from the TCGA and GTEx databases. The validation dataset was composed of 40 CRC patients who received radical surgery and 40 adjacent tissues from the specimen library of Guangzhou First People's Hospital. All patients had not received radiotherapy and chemotherapy before surgery. The format of data was converted from level 3 HTSeq-Fragments Per Kilobase Per Million (FPKM) to Transcripts Per Million (TPM) format for further analyses.
Real-Time Quantitative Reverse Transcription (qRT-PCR) analysis
Trizol reagent (Invitrogen) was used to isolate total RNA from the cells. qRT-PCR was performed according to a previous method [11]. All data were standardized with internal control GAPDH.
Functional enrichment analysis for LINC00634
Gene Set Enrichment Analysis (GSEA) has been widely used to determine whether there is an obvious difference in a set of genes between two biological states [12]. To assess the variation between LINC00634 upregulation and downregulation groups, GSEA was performed using the package ‘clusterProfiler’ in R v3.6.0 [12,13]. Based on the default settings, the process was repeated 5,000 times for each analysis, and the c2.all.v7.2.symbols.gmt (Curated) in MSigDB Collections was selected as the reference gene collection. The false discovery rate (FDR) <0.25 and adjusted p-value<0.05 were deemed as significant enrichment.
Immune cell infiltration analysis
Single-sample GSEA (ssGSEA) was conducted to analyze the immune cell infiltration of CRC by using the package ‘GSVA’ in Rv3.6.0 [14]. The RNA-seq and clinical data were retrieved from TCGA COADREAD project. All data were filtered to remove adjacent tissues and converted into Transcripts Per Million (TPM), followed by performing log2 conversion for subsequent analysis. To determine the relationship between LINC00634 and infiltration levels of 24 immune cell types, the P-value was calculated by Spearman’s rank correlation and Wilcoxon rank-sum tests.
Statistical analysis
All statistical tests were carried out with R v3.6.3. Chi-square, logistic regression, Fisher’s exact and Wilcoxon rank-sum tests were employed to assess the relationship between LINC00634 expression and clinicopathological characteristics of CRC patients. Survival package was used for the statistical analysis of survival data, survminer package was applied for visualization, and Kaplan-Meier method was employed to calculate the Disease-Specific Survival (DSS) and Overall Survival (OS) rates of CRC patients in TCGA datasets. To explore the association between clinical characteristics and OS rates, univariate and multivariate analyses were conducted using the Cox proportional hazards model. P-value of <0.05 was considered statistically significant. Finally, R language was used to draw a nomogram and establish a predictive model.
High expression of LINC00634 is related to the clinicopathological features of CRC patients
The expression levels of LINC00634 in 480 CRC tissues and 41 neighboring tissues were detected, and we observed that LINC00634 was highly expressed in CRC tissues (P<0.001, Figure 1A). At the same time, we also analyzed the expression of LINC00634 in 50 cases of CRC tissues and their matched adjacent tissues. The results demonstrated that the expression of LINC00634 was also higher in CRC tissues than in control tissues (P<0.01, Figure 1B). Due to a lack of normal tissues, the data obtained from the GTEx data were included into the control group, and the results further confirmed that LINC00634 was highly expressed in CRC tissues compared to normal tissues (P<0.01, Figure 1C). The validation dataset also achieved the same result (P<0.01, Figure 2). Collectively, these findings indicate that LINC00634 is markedly overexpressed in CRC tissues.
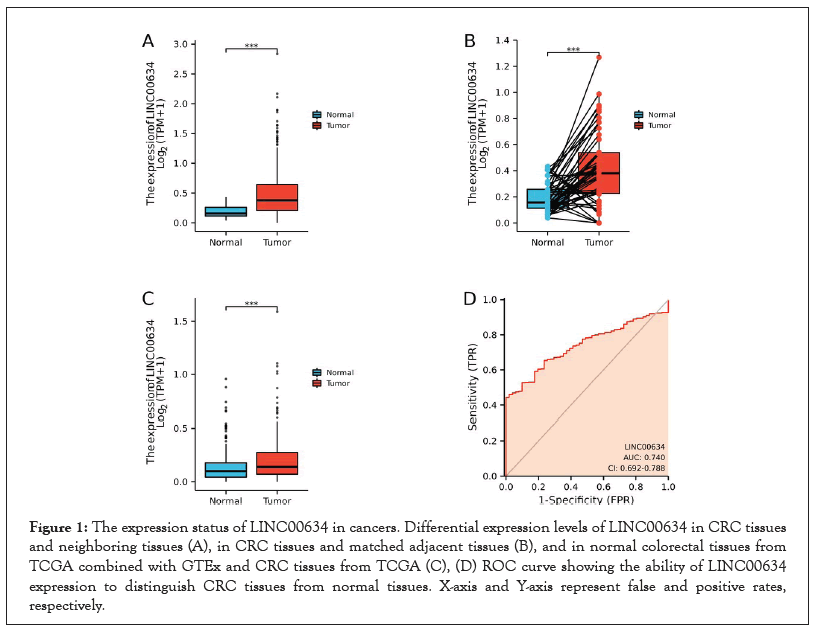
Figure 1:The expression status of LINC00634 in cancers. Differential expression levels of LINC00634 in CRC tissues and neighboring tissues (A), in CRC tissues and matched adjacent tissues (B), and in normal colorectal tissues from TCGA combined with GTEx and CRC tissues from TCGA (C), (D) ROC curve showing the ability of LINC00634 expression to distinguish CRC tissues from normal tissues. X-axis and Y-axis represent false and positive rates, respectively.
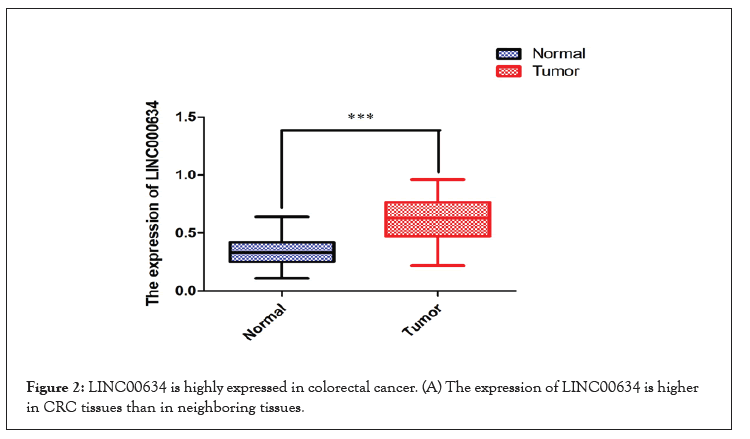
Figure 2:LINC00634 is highly expressed in colorectal cancer. (A) The expression of LINC00634 is higher in CRC tissues than in neighboring tissues.
Next, the performance of LINC00634 expression to distinguish between CRC patients and normal subjects was evaluated based on the receiver operating characteristic (ROC) curves. The Area Under Curve (AUC) of LINC00634 was 0.740, indicating that LINC00634 could serve as a good biomarker for identifying CRC and normal tissue (Figure 1D).
The clinicopathological features of CRC patients are listed in Table 1. As above mentioned, the clinical and gene expression data of 644 primary CRC patients were obtained from TCGA database. Based on the mean expression of LINC00634, the CRC patients were assigned to LINC00634 upregulation (n=322) and downregulation (n=322) groups. Subsequently, we evaluated the association between LINC00634 expression and the clinicopathological features of CRC patients. The results of Chi-square or Fisher’s exact test indicated that LINC00634 expression was associated with primary treatment outcome (P<0.05), pathologic stage (P<0.05), N stage (P<0.05), residual tumor (P<0.01) and OS event (P<0.05). Logistic regression analysis was performed to further verify the relationship between LINC00634 expression and the clinicopathological features of CRC patients. The findings demonstrated that LINC00634 expression was associated with N stage (P=0.016), pathological stage (P=0.008) and residual tumor (P=0.018) (Figure 3 and Table 2).
| Characteristic | Low expression of LINC00634 | High expression of LINC00634 | p |
|---|---|---|---|
| n | 322 | 322 | |
| T stage, n (%) | 0.714 | ||
| T1 | 8 (1.2%) | 12 (1.9%) | |
| T2 | 52 (8.1%) | 59 (9.2%) | |
| T3 | 221 (34.5%) | 215 (33.5%) | |
| T4 | 38 (5.9%) | 36 (5.6%) | |
| N stage, n (%) | 0.037 | ||
| N0 | 198 (30.9%) | 170 (26.6%) | |
| Characteristic | Low expression of LINC00634 | High expression of LINC00634 | p |
| N1 | 71 (11.1%) | 82 (12.8%) | |
| N2 | 49 (7.7%) | 70 (10.9%) | |
| M stage, n (%) | 0.177 | ||
| M0 | 243 (43.1%) | 232 (41.1%) | |
| M1 | 38 (6.7%) | 51 (9%) | |
| Pathologic stage, n (%) | 0.013 | ||
| Stage I | 52 (8.3%) | 59 (9.5%) | |
| Stage II | 137 (22%) | 101 (16.2%) | |
| Stage III | 82 (13.2%) | 102 (16.4%) | |
| Stage IV | 37 (5.9%) | 53 (8.5%) | |
| Primary therapy outcome, n (%) | 0.047 | ||
| PD | 12 (3.8%) | 21 (6.7%) | |
| SD | 4 (1.3%) | 1 (0.3%) | |
| PR | 5 (1.6%) | 11 (3.5%) | |
| CR | 139 (44.6%) | 119 (38.1%) | |
| Residual tumor, n (%) | 0.009 | ||
| R0 | 247 (48.4%) | 221 (43.3%) | |
| R1 | 4 (0.8%) | 2 (0.4%) | |
| R2 | 10 (2%) | 26 (5.1%) | |
| OS event, n (%) | 0.018 | ||
| Alive | 270 (41.9%) | 245 (38%) | |
| Dead | 52 (8.1%) | 77 (12%) |
Table 1: LINC00634 expression correlated with poor clinicopathologic features.
| Characteristics | Total(N) | Odds Ratio(OR) | P-value |
|---|---|---|---|
| T stage (T3andT4 vs. T1andT2) | 641 | 0.819 (0.556-1.203) | 0.309 |
| N stage (N1andN2 vs. N0) | 640 | 1.475 (1.077-2.024) | 0.016 |
| M stage (M1 vs. M0) | 564 | 1.406 (0.892-2.231) | 0.144 |
| Pathologic stage (Stage III and Stage IV vs. Stage I and Stage II) | 623 | 1.539 (1.120-2.118) | 0.008 |
| Primary therapy outcome (SD and PR and CR vs. PD) | 312 | 0.506 (0.233-1.053) | 0.074 |
| Age (>65 vs.<=65) | 644 | 0.859 (0.628-1.173) | 0.339 |
| Residual tumor (R1andR2 vs. R0) | 510 | 2.235 (1.166-4.472) | 0.018 |
| Perineural invasion (Yes vs. No) | 235 | 0.809 (0.447-1.455) | 0.480 |
| Lymphatic invasion (Yes vs. No) | 582 | 1.258 (0.902-1.755) | 0.176 |
Table 2: Overexpression of LINC00634 predicts poor prognosis in colorectal cancer patients.
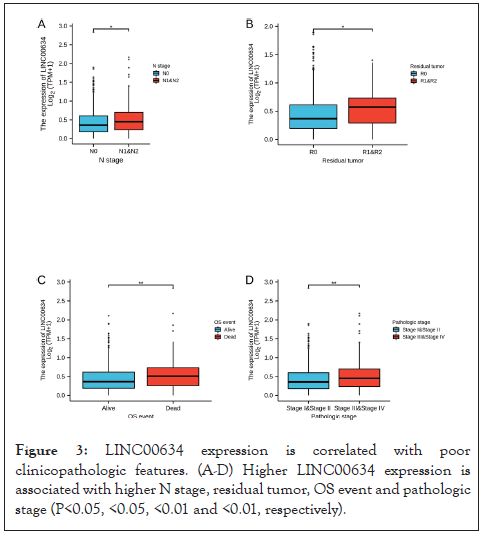
Figure 3: LINC00634 expression is correlated with poor clinicopathologic features. (A-D) Higher LINC00634 expression is associated with higher N stage, residual tumor, OS event and pathologic stage (P<0.05, <0.05, <0.01 and <0.01, respectively).
LINC00634 expression is related to poor prognosis in CRC patients
Kaplan-Meier survival curve was applied to determine the correlation between LINC00634 expression and the OS/DSS of CRC patients. The results indicated that LINC00634 expression was positively associated with poor OS and DSS in CRC patients (P<0.01, Figure 4A and P=0.008, Figure 4B, respectively).
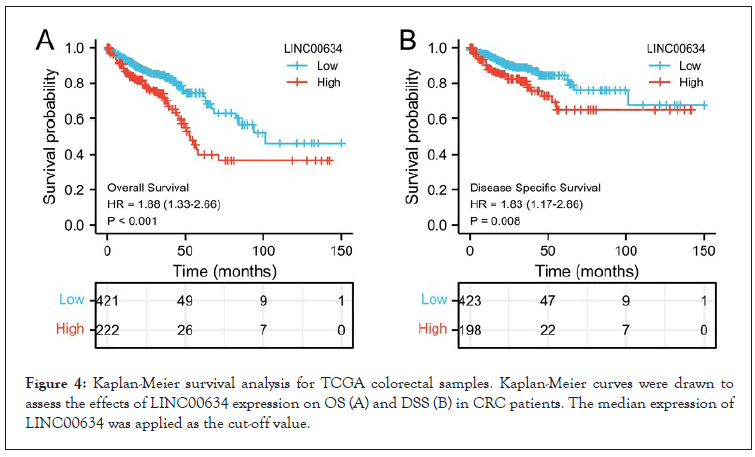
Figure 4: Kaplan-Meier survival analysis for TCGA colorectal samples. Kaplan-Meier curves were drawn to assess the effects of LINC00634 expression on OS (A) and DSS (B) in CRC patients. The median expression of LINC00634 was applied as the cut-off value.
Identification of prognostic factors for OS in CRC
The Cox univariate and multivariate analyses of prognostic factors for OS in CRC patients are revealed in Table 3. Notably, the significant univariate variables were age (P<0.01), LINC00634 (P=0.039), T/N/M stage (P<0.01) and residual tumor (P<0.001). Multivariate analysis further showed that age (P=0.002), LINC00634 (P=0.019), T stage (P<0.05), M stage (P<0.05) and residual tumor (P<0.05) were independent prognostic factors for OS in CRC patients. Based on Cox regression analyses, a nomogram was constructed for internal verification, and a predictive model was established (Figure 5). In internal verification, the c-index of the nomogram was 0.772 (95% CI: 0.744-0.799).
| Characteristics | Total (N) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P-value | Hazard ratio (95% CI) | P-value | ||
| T stage (T3andT4 vs. T1andT2) | 640 | 2.468 (1.327-4.589) | 0.004 | 3.610 (1.284-10.147) | 0.015 |
| N stage (N1andN2 vs. N0) | 639 | and | <0.001 | 1.569 (0.899-2.739) | 0.113 |
| M stage (M1 vs. M0) | 563 | 3.989 (2.684-5.929) | <0.001 | 2.206 (1.187-4.101) | 0.012 |
| Residual tumor (R1 and R2 vs. R0) | 509 | 4.609 (2.804-7.577) | <0.001 | 2.075 (1.133-3.801) | 0.018 |
| Gender (Male vs. Female) | 643 | 1.054 (0.744-1.491) | 0.769 | ||
| Race (Black or African American and White vs. Asian) | 394 | 0.840 (0.205-3.438) | 0.809 | ||
| Characteristics | Total (N) | Univariate analysis | Multivariate analysis | ||
| Hazard ratio (95% CI) | P-value | Hazard ratio (95% CI) | P-value | ||
| LINC00634 (High vs. Low) | 643 | 1.450 (1.019-2.063) | 0.039 | 1.736 (1.094-2.755) | 0.019 |
| Age (>65 vs.<=65) | 643 | 1.939 (1.320-2.849) | <0.001 | 2.213 (1.332-3.675) | 0.002 |
Table 3: Univariate and multivariate analysis for LINC00634.
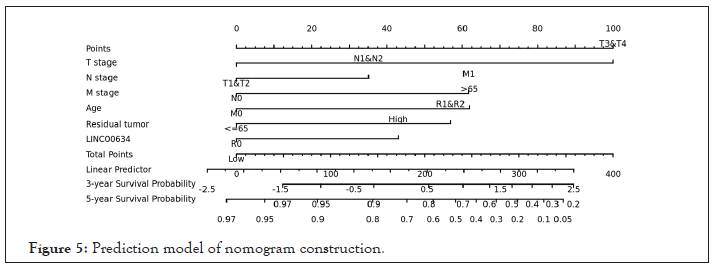
Figure 5: Prediction model of nomogram construction.
LINC00634 expression and its significantly involved functions
GSEA was carried out to explore the biological functions related to LINC00634 expression.There were 55 datasets with significantly differential enrichments in LINC00634 upreglation group, and we identified the target 6 datasets with high Normalized Enrichment Score (NES). Most of these pathways were associated with gene transcription and gene methylation (Figure 6).
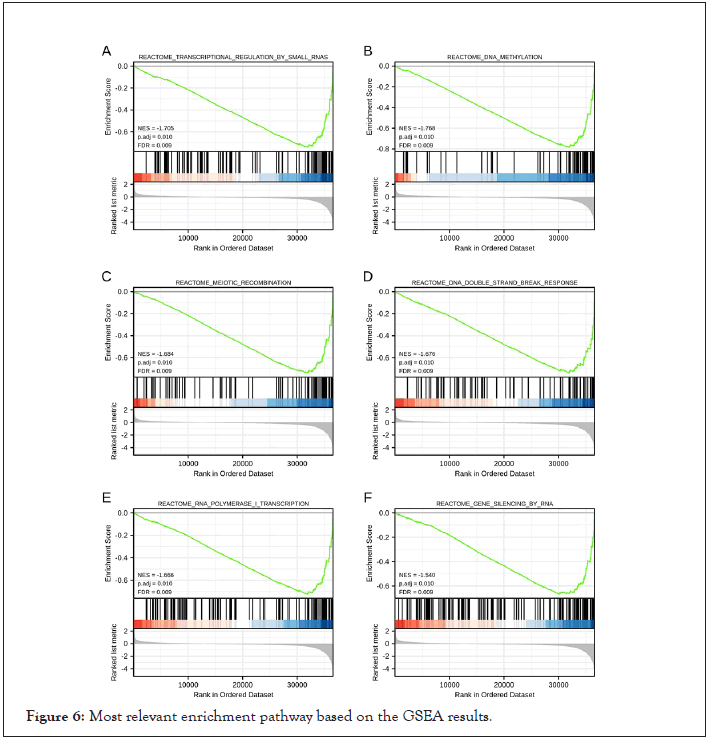
Figure 6: Most relevant enrichment pathway based on the GSEA results.
LINC00634 expression and immune cell infiltration
The relationship between LINC00634 expression and immune cell infiltration was examined by ssGSEA (Figure 7A).
Our findings demonstrated that LINC00634 expression was negatively associated with the infiltration levels of T central memory (Tcm) cells (P<0.01, Figure 7B-C), while positively associated with the infiltration level of natural killer (NK) CD56 bright cells (P<0.01, Figure 7D-E).
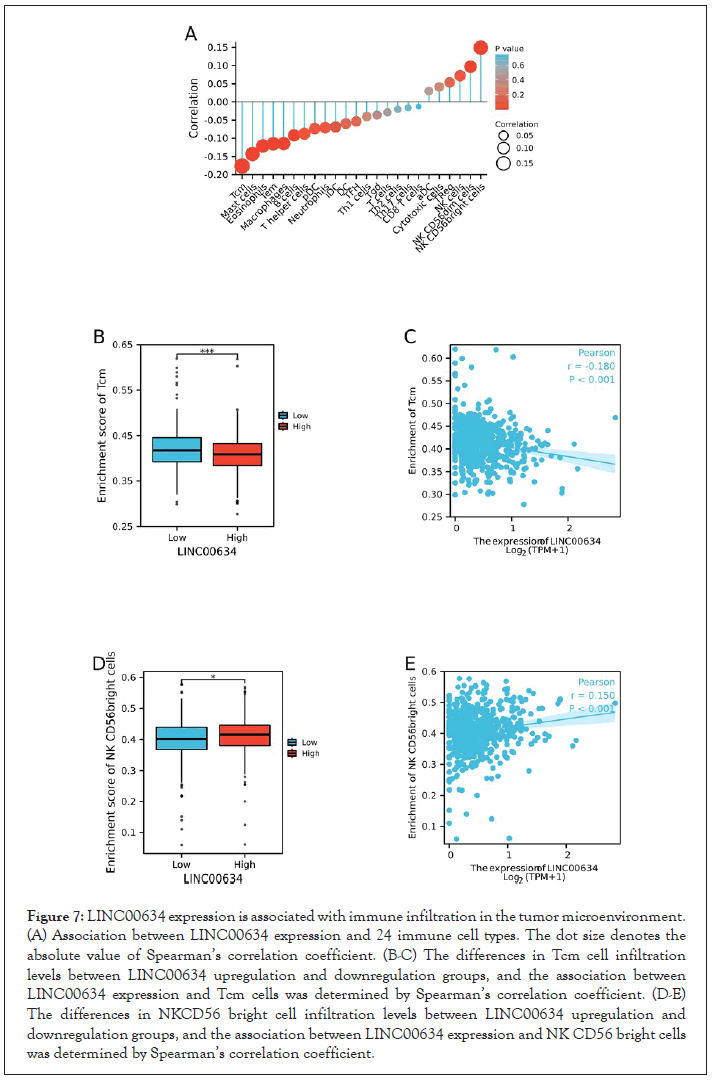
Figure 7: LINC00634 expression is associated with immune infiltration in the tumor microenvironment. (A) Association between LINC00634 expression and 24 immune cell types. The dot size denotes the absolute value of Spearman’s correlation coefficient. (B-C) The differences in Tcm cell infiltration levels between LINC00634 upregulation and downregulation groups, and the association between LINC00634 expression and Tcm cells was determined by Spearman’s correlation coefficient. (D-E) The differences in NKCD56 bright cell infiltration levels between LINC00634 upregulation and downregulation groups, and the association between LINC00634 expression and NK CD56 bright cells was determined by Spearman’s correlation coefficient.
In this study, we sought to determine the expression of LINC00634 in CRC and its association with the diagnosis and prognosis of CRC. Thus far, there are only few studies about the relationship between LINC00634 and tumors. A recent study found that LINC00634 was upregulated in esophageal cancer tissues, and its expression was related to the TNM staging of esophageal cancer patients. Mechanistically, LINC00634 regulated the downstream target gene BcI2LI by sponging miR-608 to attenuate inhibition, promoted the proliferation of esophageal cancer cells and inhibited cells apoptosis [10]. Our results indicated that LINC00634 was highly expressed in CRC tissue, and was related to the poor prognosis of the patients. Many studies have reported the key roles of DNA methylation and RNA polymerase I in human cancers [15,16]. Here, we also focused on predicting the potential mechanism of LINC00634 in regulating the occurrence of CRC. GSEA functional enrichment results showed that LINC00634 was involved in the processes of DNA methylation and RNA polymerase I transcription, and affected gene expression at the transcription level. The aforementioned results corroborated that LINC00634 is abnormally expressed in CRC, and may affect the progression of CRC by implicating in the processes of DNA methylation and RNA polymerase I transcription.
An extensive body of research showed that LINCRNAs are becoming a key molecule of immune regulation [17-20], and play functional roles in regulating cancer immunity and Tumor Immune Microenvironment (TME) [21]. Furthermore, the correlation between LINCRNAs and immune cell infiltrate has also been explored in certain cancers, which implies the potential of LINCRNAs in evaluating the immune cell infiltrate of tumor [22,23]. Another purpose of this study was to explore the association between LINC00634 expression and the infiltration levels of various immune cells in CRC. Our results demonstrated a correlation between LINC00634 expression and the infiltration levels of Tcm and NK CD56 bright cells in CRC. Moreover, LINC00634 could inhibit the functions of Tcm and promote the function of NK CD56 bright cells, thereby exerting its potential functions on CRC. Cox regression analysis revealed that LINC00634 expression, T stage, M stage, residual tumor and age were independent prognostic factors for colorectal carcinoma. Interestingly, Kaplan-Meier survival analysis showed that patients with LINC00634 upregulation exhibited lower OS and DSS than those with LINC00634 downregulation. Based on the results of Cox regression analysis, we drew a nomogram and performed internal verification in order to have a more accurate prediction of the 3-year and 5-year survival rates of colorectal carcinoma patients, and this model had a certain practical significance for the development of clinical treatment. In internal verification, the c-index of the nomogram was 0.772 (95% CI: 0.744-0.799). Therefore, the prediction model had good accuracy. However, there are some inevitable limitations that need to be addressed. First, the current research data was mainly obtained from public datasets, and the results of bioinformatics analysis should be further validated through experimental research. Second, it is necessary to obtain data from different cohorts in order to improve the credibility of this study. Finally, retrospective research still has many shortcomings especially the accuracy of data and lack of some information. Therefore, future experimental research is warranted to validate our findings.
In summary, LINC00634 was increased in CRC tissues, and its expression was related to poor OS and DSS. Moreover, LINC00634 was involved in the development of CRC by affecting gene transcription levels and immune infiltrating cell functions. Our findings indicate that LINC00634 can serve as a promising biomarker for the diagnosis and prognosis of CRC.
Acknowledgements
We acknowledge TCGA and GTEx databases for the free use of gene expression and clinical datasets.
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Ethics approval and consent to participate
This research was reviewed and approved by The Ethics Committee of Guangzhou First People’s Hospital (No. K-2018-140-04). Written informed consent was obtained from each patient involved in the study.
Data availability statements
The data that support the findings of this study are openly available in [TCGA (https://portal.gdc.cancer.gov/) COADREAD project] and [GTEx(https://www.gtexportal.org/)].
Consent for publication
Not applicable.
Author contributions
LF organized, wrote and critically modified the manuscript; NYQ modified the manuscript; FFYH drafted the manuscript and was responsible for the acquisition of data; LF participated in the data analysis; LF contributed to the literature search. All authors read and approved the manuscript, and agree to be accountable for all aspects of the research and manuscript.
Funding
This work was supported by the grants from the National Natural Science Foundation of China (81871905).
Citation: Liu F, Fu F, Nie Y (2021) Bioinformatic Analysis: The Role of LINC00634 in Colorectal Carcinoma. J Clin Trials. 11:481.
Received: 06-Sep-2021 Accepted: 20-Sep-2021 Published: 27-Sep-2021 , DOI: 10.35248/2167-0870.21.11.481
Copyright: © 2021 Liu F, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.