Journal of Plant Biochemistry & Physiology
Open Access
ISSN: 2329-9029
ISSN: 2329-9029
Research Article - (2019)Volume 7, Issue 2
Objective: The goal was to determine the correlation of anythocyanin production and receptor expression in different Tangela (Monjara tangelo) tissues to their color and known growth rates, and pin the molecular factors underlying vine whipping and defensive compound production.
Methodology: Monjara tangela were exposed to varying levels of light:dark in regulated growth chambers for a period of eight weeks. Young vines and root scrapings were taken at each day in the experiment for cDNA extraction and quantitative PCR performed for chlorophyll, xanthophyll, and anthocyanin receptor expression.
Results: Growth of finely glaucous vines was constant throughout the study period, with consistent development of fibrous swellings along the root apical meristem. In all conditions, flowering did not occur, with asexual runners produced consisting of no bracts and very few true leaves. Between the 3th and 5th week, the root meristem began to accumulate anthocyanin and turned red while the apical meristems differentiated into whip primordia. Chlorophyll a accumulation ceased at the onset of anthocyanin synthesis and the protein content per unit leaf area declined. Western blot analysis confirmed that reduced chlorophyll a and b expression at this time correlates with the maturation of the roots and the reallocation of proteomic resources towards developing the defensive semichemicals associated with its soporiforous spore production. The data can be used for future gene knockout experiments and better management of plant cultivation.
Chemical ecology; Botany; Plant physiology; Plant biochemistry
Commercial and academic research into the primary and secondary metabolites of Poaceae-type organisms has led to considerable gains in our knowledge of plant chemical defenses and their potential applications. Unfortunately, few have studied related non-Poaceae crops and non-crop organisms [1]. Among the first generation of model organisms is the eudicotic Monjara tangela (Vitaceae), commonly known as Tangela and the pre-evolutionary form of Monjara tangrowth (likely a synonym) [2]. Known for its constant and rapid vine and shoot development, its runners snap off easily if grabbed and are infamous for entangling hikers in the forests of Chiba Prefecture, Japan, where it is endemic [3]. The various mono- and dikaryotic spores from Tangela and other relatives have been investigated for their soporific, toxic, and paralytic abilities. The implications for these compounds in the pharmaceutical and combat arenas are well documented [1,4].
Still unanswered are some of the ontogenetic and developmental level questions surrounding the persistent vine growth. Transcriptomics analysis of the distantly related Sunflora kernes suggested that whipping behavior is coded in differential gene expression leading to rapid cell division of the apical meristems [5], while electrochemical assays with Rafflesia vileplume demonstrated remarkable re-location of water resources through the plants’ xylem tubes, leading to sustained turgor pressure at the vine tip [6]. Yet these vine whipping instances are specifically triggered by training and by attacks from herbivorous insects or other ectobiotic antagonists, and do not compare with the sustained growth seen in Tangela [2]. As such, the subject should be seen as a distinct system meriting further exploration.
The nutritional and nutraceutical markets for M. tangela are nascent but show great promise [7,8]. That alone would motivate research into the expression of phytochemicals such as alkaloids and flavonoids in this and any other Poaceae-type, but the high probability that chemical expression correlates with vine development suggests this research could solve the consistent growth riddle as well as providing applicable data for industry. Anthocyanin in particular is thought to solve the question originally posed by Applebaum and Natochenny [9], namely that of the uncharacteristic blue tinge of M. tangela tissue relative to other terrestrial plants, more similar to that of seaweeds than Vitaceae. The link to anthocyanin comes from the dark blues found in certain varieties of grape (Vitus, Vitaceae). We hypothesize that the color in M. tangela is due to hyperactive production of anthocyanin in the relevant tissues, and, based on prior research on vine whipping [5,8], we go further to claim that this overexpression correlates with the frenzied growth iconic to this genus, in part inspired by the landmark studies by Willow et al. on Tangela razor leaf-related gene expression and stun spore microstructures [10,11].
This manuscript describes work into the transcriptomics of M. tangela , with a focus on anthocyanin production, detection, reception, and membrane translocation genes. The study measured how this and other flavonoids such as chlorophyll a and were distributed among the organism’s main tissue types: the blue-green vines, the red taproots, and the black ocular nidus. This study is novel, far-reaching, and likely to springboard future studies of non-crop Monstrasinu research.
A previous paper by Oak and Oak describes the then de novo assembly and annotation of Tangela’s transcriptome and production of a micro-array for key genes [12]. Commercially available M. tangela microarrays were used as per manufacturer’s instructions (GARraY Inc.) and with the appropriate microarray analysis software [12,13], optimized as per the published protocol [12]. Peri-domestic Tangela were obtained from tall grass near the Celadon City gymnasium and replicates of n=4 were incubated in laboratory growth chambers for three months at the following light:dark cycles: 12:12, 6:18, 18:6, and 24:0. Culturing protocols have been optimized in our lab for some time and were previously published [9]. Weekly collections from wild specimens were considered, but ultimately not collected due to the weather as an uncontrollable variable. At weekly intervals vine growth and changes in overall mass and water consumption were measured. Samples were taken for RNA extraction using sterile razors, sampling from the apical, medial, lateral, stem [main shoot], and root vines. Tissue was stored in RNAlax at -80°C until the end of the experiment, when RNA was extracted as per the chlorophyll-phenol protocol [14] and cDNA produced using a Stunfisk® Reverse Transcriptase kit as per manufacturer instructions [12]. Pooled samples from each subject were combined but separated per tissue type. Transcriptome analysis was done by the Celadon University Genome Core Facility using an Illumise® Microarray Plate Reader. The raw SRA data has been stored in GenBank (Accession Number LT4815162342).
At the end of the study, 1 mg cuttings from the respective tissues were taken and their anthocyanin and chlorophyll a and b levels analyzed using a GC-MS and a Reynolds HCl test respectively [15,16]. As to file for plant flavonoids was generated from hits in the UNIPROT database of proteins from the related pocket monsters [17]. Efforts to generate text from SCIgen failed to pass the target journals anti-plagiarism detectors and had to be revised [8] but we are otherwise confident a study about a pocket monster can be published, as this invited article is in a predatory journal that likely does not practice peer-review [18]. Statistical analysis was done with an ANOVA using R [19]. Following the end of the experiment, the Tangela specimens were maintained in the Celadon University botanical gardens pending IPCUC approval for re-release to the peridomestic environment where they were found.
Growth performance across the different light-dark cycles did not show statistically significant variance (p>0.1), nor were there any significant effects of light periodicity on the expression of anthocyanin genes on the microarrays. Expression of papaverine, phyto-opiate, and linalool genes was significantly upregulated with decreased dark cycles below 12 hours (p<0.01), with no difference between the 18:6 and 24:0 L:D samples. This expression correlated with observed increased production of resting spores by all specimens in such chambers following a continuous 20-22 hours of light, which suggests an autosomnolent behavior.
Anthocyanin production did not differ significantly with periodicity or with growth rates, the latter of which were constant anyway, but expression values for anthocyanin receptors (Table 1) were differentially expressed across the tissues and were significantly higher than those of typical plants.
| Anthocyanin Type | Apical Vine | Medial Vine | Lateral Vine | Stem Vine | Root Vine |
|---|---|---|---|---|---|
| Aurantinidin | 550 ± 2 | 0 | 3252 ± 22 | 0 | 30 ± 1 |
| Cyanidin | 223 ± 4 | 0 | 0 | 0 | 445 ± 4 |
| Delphinidin | 0 | 0 | 0 | 122 ± 3 | 2233 ± 22 |
| Europinidin | 42234 ± 490 | 12288 ± 777 | 44422 ± 3 | 32 ± 4 | 200 ± 3 |
| Malvidin | 0 | 0 | 0 | 0 | 0 |
| Peonidin | 0 | 0 | 0 | 12 ± 1 | 0 |
| Petunidin | 0 | 4312 ± 1 | 3242 ± 34 | 12 ± 3 | 0 |
| Rosinidin | 0 | 0 | 3422 ± 70 | 0 | 10 ± 0 |
Table 1: Differential expression of anthocyanin receptors across Tangela vines. Expression in RPKB-Reads per thousand base pairs.
Anthocyanin content varied significantly in the different vines, leading to the overall distribution as summarized in Figure 1.
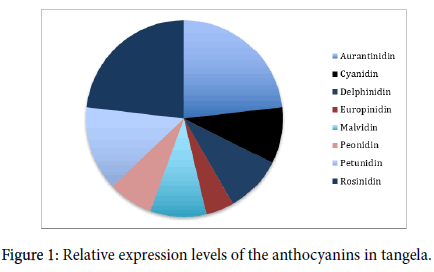
Figure 1: Relative expression levels of the anthocyanins in tangela.
Aurantinidin and Rosinidin dominate, while Europindin levels were the lowest. Chlorophyll was not detected in any sample with the Reynolds test, but this is not due to absence or decay of chlorophyll in the pooled, dead tissue samples, but a mechanical failure of the laboratory’s green wavelength emitter that unfortunately could not be resolved in time for publication.
Observed anthocyanin receptor differential expression in different vines suggests a significant correlation between the color and growth rates of these vines over the lifetime of a Tangela, consistent regardless of periodicity and seasonality [20]. This would enable year-long growth and development ideal for the needs of the organism, as has been found in the Indonesian daikon radish Oddium wanderus [21]. This differential receptor expression responds to the otherwise evenly circulating anthocyanin levels in the phloem to produce the coloration noted in typical Tangela [22,23]. Future research will need to look at anthocyanin levels in the ‘shiny’ variants and see how they compare to what we observed here [24].
Anthocyanins can drive root and stem apical meristerm growth and differentiation into new tissue through a multitude of ways. The Toll immune pathway uses malvidin and europinidin as transcription modulating factors, prior to differentiation of root tissues in Ligustrum sp. [25]. Malvidin and peonidin, and rarely rosinidin, are anthocyanins whose role in leaf stoma opening and closing genes has been explored in depth in M. tangela [4] and the data conforms with the accepted findings both in specific (Lilium bella , Nymphaeae saltadus , Lera garlandus) and non-specific grass-type organism research [26,27].
The high expression of receptors for europinidin and moderately high expression of receptors for petunidin and rosinidin in the lateral vines; the extremely high expression of receptors for europidin and moderately high expression of petunidin receptors in medial vines; and the extremely high expression of europidin in the apical vines (Table 1); all combined with the relatively low levels of europinidin in Tangela overall, suggest that europinidin above all other anthocyanins is mobilized to the vines involved in vine whipping, grass knots, and frenzied growth, whose abundant receptors for it suggest it is a transcription or signaling factor stimulating cell division and differentiation. The data also suggests that delphinidin is involved in the shoe-like structure of the taproot, as was remarked for the egg palm Cocos ovulum.
The putative medicinal properties of these anthocyanins also suggest that certain tissues of Tangela could be used in extractions to produce commercial quantities of these compounds, though whether teas or tisanes of Tangela vine would have any therapeutic effect is beyond the scope of this work, though others are exploring it [28-30]. Certainly, extraction of the anthocyanins as dyes or colorants is possible.
This paper not only defines and delineates the anthocyanin types present in the various tissues of M. tangela , but also identified where these anthocyanins are being received by the cells, meaning where their activity is highest and most likely correlated with the organism’s behavior and development. Further development of metagenomic data for Tangela and other Poaceae-like organisms is anticipated, with an eye towards commercial applications.
The authors would like to thank Celadon City Genome Core Facility and its staff. Funding was provided by Kanto Regional Development Grant, PKE-114.
Citation: Applebaum E, Schlangemann H, Shelomi M (2019) Anthocyanin Receptor Expressions Across Tangela (Monjara tangela) Vines. J Plant Biochem Physiol 7: 237. doi: 10.35248/2329-9029.19.7.237
Received: 17-May-2019 Accepted: 17-Jun-2019 Published: 24-Jun-2019 , DOI: 10.35248/2329-9029.19.7.237
Copyright: © 2019 Applebaum E, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.