Journal of Cell Science & Therapy
Open Access
ISSN: 2157-7013
ISSN: 2157-7013
Mini Review - (2021)Volume 12, Issue 6
End resection is a central step in DNA homologous recombination (HR) and the HR-mediated repair of DNA double-strand breaks (DSBs), which degrades 5’-end strands at DSBs by several kilo-bases and thus creates 3’-end single-strand DNA overhangs. A critical long-standing question is how the 3’-end strands are protected during end resection. Now, this question is answered. Liu et al. found that the protection of 3’-end strands is achieved by the formation of an RNA-DNA hybrid. RNA polymerase III is responsible for catalyzing the RNA strand in the hybrid. Thus, RNA polymerase III is an essential factor for HR, and the RNA-DNA hybrid is an essential HR intermediate.
DNA homologous recombination; RNA polymerase III; RNA-DNA hybrids
DNA Homologous Recombination (HR), a ubiquitous basic biological process, plays an essential role in cell growth, gamete production, genome diversity, and evolution of species [1,2]. It is also crucial in maintaining genomic integrity because it is required for the repair of DNA double-strand breaks (DSBs) [3]. In humans, the defects of HR cause cancers, neurodegenerative diseases, and aging [4,5]. Thus, a thorough elucidation of an HR process at the molecular level can not only advance the understanding of basic DNA metabolism but also promote the development of relevant drugs for treating cancers and neurodegenerative diseases.
In the 1930s, the phenomena of recombination of genetic materials were observed [2], and in 1944, the genetic materials were identified to be DNA [6]. Since then, HR has been extensively studied for nearly 80 years, and a great progress has been made in understanding the molecular process of an HR event [1,2,7]. Based on the current model, HR comprises three major steps: end resection, strand invasion, and resolution of Holliday junctions [5]. End resection involves removing a few kilobases from the 5’-end strand at DNA DSBs but keeping the 3’-end strand intact [8]. Next, RAD51 binds to the 3’-end singlestrand DNA (ssDNA) strand to generate a nucleofilament [1]. This nucleofilament invades a homologous DNA molecule (often a sister chromatid) and acts as a primer for subsequent DNA pol δ-mediated DNA synthesis, resulting in the formation of a Holliday junction [9]. Finally, this Holliday junction is resolved by the nucleases Mus81-Eme1 [10], GIN1 [11], and SLX4 [12]. Although a frame of the HR process is established, numerous critical questions remain unanswered. For example, the basic mechanism for protecting the 3’-end strand during end resection was not known. Previous studies suggested that RPA (replication protein A, a single-strand DNA binding protein in eukaryotes) binding might protect the 3’-end strands from the digestion by Dna2 or other nucleases during end resection [13,14]. However, this suggestion or hypothesis lacked solid experimental evidence. In addition, it is very unlikely that RPA binding protects the 3’-end strand because of the following three principal reasons. First, cells do not have a mechanism that ensures that RPA binding to the 3’-end ssDNA strand is certainly prior to nuclease attack. Second, cells do not have a mechanism to guarantee that every nucleotide on a 3’-end strand of several kilobases in length is bound by RPA. Third, RPA binding to ssDNA, as other proteins interact with dsDNA or ssDNA, frequently dissociates, and the dissociation leaves one or several regions of the 3’-end ssDNA strand exposed to nuclease attack. More critically, in fission yeast RPA binding stimulates Dna2 digestion of ssDNA [15].
In the last several decades, astonishing progress was the identification of a number of protein factors directly involved in the HR process, including the MRN (MRE11-RAD50-NBS1) complex [16,17], CtIP [18], DNA2 and EXO1 [19,20], BRCA1/2 [21,22], RAD51 [23,24], RAD52 [25,26], RPA [27], DNA helicase BLM [28-30], histone remodeling factors (INO80 [31], RNF8/168 [32,33]), SLX4 [12], GEN1 [11], and so forth. These factors are either required for end resection or directly participate in strand invasion and resolution of Holliday junctions. In mammalian cells, although mutations on the majority of these factors do not cause cell death, their defects result in severe genomic instability and predispose to a variety of carcinogenesis [4,5]. In addition, HR defects also cause severe developmental disorders [34].
Recently, RNA polymerase III (RNAPIII) was identified as an essential factor for HR in human cells [35]. RNAPIII was demonstrated to catalyze RNA synthesis at DSBs. This RNAPIIIcatalyzed RNA strand pairs with the 3’-end ssDNA strand to form an RNA-DNA hybrid, protecting the 3’-end ssDNA strand during end resection (Figure 1). Thus, a long-standing crucial question of how the 3’-end ssDNA strand is protected during end resection was finally resolved. In more detail, RNAPIII is recruited to DSBs through a specific interaction between the MRE11 subunit of the MRN complex and the specific subunits RPC4 and RPC6 of RNAPIII. It is independent of the cell cycle phase that RNAPIII is recruited to DSBs, but RNA synthesis takes place only in the S/G2 phase. When the RNA synthesis is dysfunctional either by a reduced level of RNAPIII or by inhibition of RNAPIII activity, the rate of HR correspondingly decreases. Moreover, as expected, genetic deletion significantly increases when the RNAPIII-mediated RNA synthesis is disrupted. Thus, RNAPIII is a newly uncovered essential factor for HR and HR-mediated repair of DSBs, and the RNA–DNA hybrid is an essential intermediate.
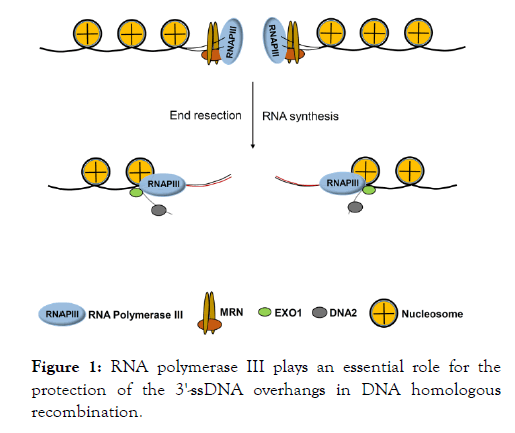
Figure 1: RNA polymerase III plays an essential role for the protection of the 3'-ssDNA overhangs in DNA homologous recombination.
Next, an impending question is how the RNA strand in the RNA–DNA hybrids is removed. Logically, an RNA helicase, or an RNA nuclease, or a combination of them, should be involved in removing the RNA strand. A biochemical approach, together with genetics, can identify the enzymes responsible for digesting or removing the RNA strand. The discovery of the RNA–DNA hybrid intermediate may also promote the solution of some other long-standing questions, such as what are the exact biochemical actions of BRCA1, BRCA2, and RAD52 in HR. Furthermore, it is also highly anticipated that a thorough elucidation of the HR process, including the mechanism of removing the RNA strand, should provide new avenues to develop drugs against some types of cancers.
We thank all members of the Kong Lab for discussion and support. This work was supported by grants from the National Natural Science Foundation of China (no. 31230021), the Ministry of Science and Technology of China (2013CB911000), the Peking-Tsinghua Center for Life Sciences, the National Key Laboratory of Protein and Plant Gene Research, and the Scientific and Technological Innovation Program of Higher Education Institutions in Shanxi (2020L0497).
Citation: Liu S, Liu X, Kong D (2021) A Recently Discovered Essential Factor for DNA Homologous Recombination: RNA Polymerase III. J Cell Sci Therapy. 12:302.
Received: 01-Jun-2021 Accepted: 15-Jun-2021 Published: 22-Jun-2021 , DOI: 10.35248/2157-7013.21.12.302
Copyright: © 2021 Liu S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.