Transcriptomics: Open Access
Open Access
ISSN: 2329-8936
ISSN: 2329-8936
Research Article - (2022)Volume 8, Issue 4
As of July 25-2020, 643,412 people in more than 215 countries have been victims of the new type of coronavirus, SARS-CoV-2. Thereby, there is a huge effort to develop a strategy to treat, and or prevent people from SARS-CoV-2 infection. Those efforts could be mainly categorized as drug repurposing, anti-SARS-CoV-2 antibodies from people who recovered, and vaccines. However, there is currently no specific treatment available against SARS-CoV-2 infected patients. That`s why many new approaches and ideas are still studied every day for the treatment of SARS-CoV-2 infected patients. Antisense therapy is one of these promising approaches to target SARS-CoV-2 genomic RNA specifically and inhibit its activity upon incorrect viral RNA processing. In this study, Antisense Oligonucleotide (ASO) candidates targeting SARS-CoV-2 genomic RNA were designed. High-scored ASOs with a high potential to inhibit SARS-CoV-2 replication and transcription by inducing cleavage of the viral genomic were determined among ASO candidates. For the future, those promising ASOs can be synthesized followed by required modifications and test on SARS-CoV-2 infected Vero cells to screen their efficacy for the treatment of SARS-CoV-2 infected patients.
Antisense oligonucleotides (ASOs); Antisense RNA therapy; Drug development; SARS-CoV-2
SARS-CoV-2: Severe Acute Respiratory Syndrome Coronavirus-2; ACE2: Angiotensin-Converting Enzyme 2; ASOs: Antisense Oligonucleotides; 5Me-Dc: 5-Methyl Deoxycytosine; 7-deaza-dG: deaza G; 2'-O-MOERNA: 2'-O-Methoxy-Ethyl 5me Uridine; PS: Phosphorothioate Linkage.
SARS-CoV-2 (severe acute respiratory syndrome coronavirus-2) is a type of single-stranded RNA virus that infects mammals and birds. SARS-CoV-2 utilizes spike (S) proteins in the outer surface to attach host cells through the angiotensin-converting enzyme 2 (ACE2) receptors which mainly on the surface of epithelial cells of the pulmonary alveolus. On the virus side, the RBD domain located on Spike (S) protein is responsible for the host attachment [1-3]. Once the viral attachment occurs, viruses enter into the host cells, following endocytosis pathways or direct fusion. The viral RNA genome is then released in the cytoplasm. Positive-sense single-stranded viral genome is then directly translated by the host ribosomes without any additional modifications or process. At this point, viral particles are formed through viral RNA translation, followed by a new virion assembly [4-6]. COVID-19 caused by SARS-CoV-2 infection has affected over 15,971,347 people and killed 643,412 in more than 215 countries as of July 25, 2020 [7]. Therefore, there is an urgent need for a relevant treatment strategy. However, there is currently no specific treatment available against SARS-CoV-2 infected patients. For this purpose, several repurposing antiviral drugs such as remdesivir, favipiravir, hydroxychloroquine, and lopinavir have been applied around the world to treat SARS-CoV-2 infected patients [8-10]. However, drug repurposing studies are a short-term effort to fulfill an urgent need. That`s why it cannot help find effective treatments for the SARSCoV- 2 infection. Many efforts have been also directed to develop anti-SARS-CoV-2 antibodies from people who recovered to treat those patients. Besides, many institutions and companies have a very big effort to develop efficient vaccines to provide protective immunity to the world population as well [9,11]. On the other site, antisense therapy could be a choice to specifically treat SARS-CoV-2 infection among other treatment strategies; vaccine, antiviral drug repurposing, and anti-SARS2-CoV-2 antibody approach [9,12]. In this case, Antisense Oligonucleotides (ASOs) targeting specific sequences on mRNAs, small RNAs, or long non-coding RNAs enter the cell following an unclear mechanism. They recognize and hybridize specifically to targets based on Watson-Crick base pairing which results in the hetero-duplex formation (ASO/target RNA). At this point, hetero-duplex structures lead to suppression of translation through either by preventing ribosomal assembly at the 5' cap directly or cleavage by RNase H (Figure 1) [12,13-18]. For instance, miravirsen (SPC3649) is an experimental antisense drug used for the treatment of hepatitis C by targeting human miRNA, miR-122 which protects HCV RNA against nuclease degradation, and 15mer ASO is already in Phase II clinical trials as of 2017 [19,20]. Such studies on antisense therapy are accumulating and will be developed further for the treatment of viral infections in the future with the help of its low toxicity, high specificity, and low production cost [11]. In the case of SARS-CoV-2 treatment, SARS-CoV-2 genomic RNA can be targeted by ASOs and block the translation of viral particles upon incorrect viral RNA processing. Therefore, antisense therapy could be also a promising approach to treat viral infections specifically through the inhibition of viral genome activity [12].
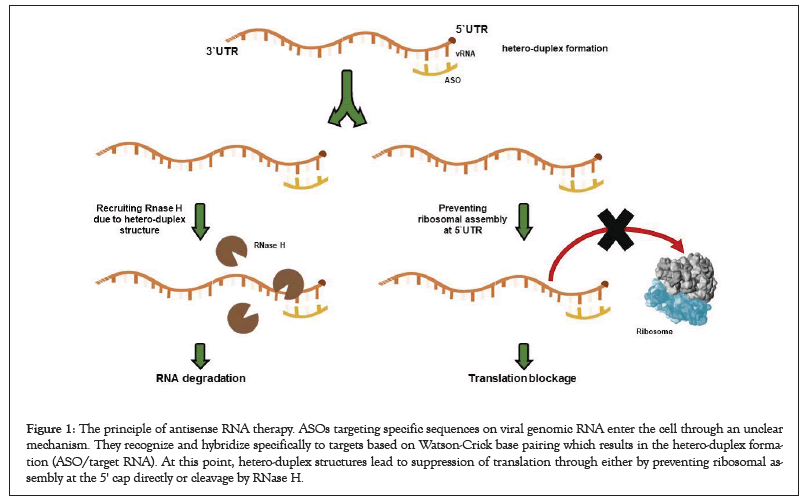
Figure 1: The principle of antisense RNA therapy. ASOs targeting specific sequences on viral genomic RNA enter the cell through an unclear mechanism. They recognize and hybridize specifically to targets based on Watson-Crick base pairing which results in the hetero-duplex formation (ASO/target RNA). At this point, hetero-duplex structures lead to suppression of translation through either by preventing ribosomal assembly at the 5' cap directly or cleavage by RNase H.
In this study, I aimed to design promising ASOs targeting SARSCoV- 2 to inhibit viral genome replication and transcription. Studies on fomivirsen, mipomersen and miravirsen, antisense drugs, were taken as reference works to design this idea. To this purpose, 5`UTR, 3`UTR, and start codon in the SARS-CoV-2 genomic RNA were analyzed to find out the regions without secondary structures. ASOs were then designed by targeting those available regions. All ASO candidates were evaluated through in vitro and in vivo tested parameters by Aartsma-Rus et al. [13] and screened for off-targets. In the final stage, modifications required for functionality were determined for each promising ASOs.
ASO design
The reference sequence of SARS-CoV-2 genomic RNA (GenBank: MT385474) was used to design ASO candidates (NCBI, 2020). The design was primarily based on targeting 5`UTR, 3`UTR, and start codon (Figure 2). In particular, ASO candidates were targeting an open region in the secondary structure of the SARSCoV- 2 genomic RNA as predicted by m-fold for each targeting site. ASO candidates were scored through the parameters as described by Aartsma-Rus et al. [13] previously. In this purpose, Tm values, ASO length, molecular weight, number, and percentage of GCs nucleotides, secondary structure formation, and dimer formation were calculated with the Oligonucleotide Properties Calculator tool [21]. All ASO candidates were scored based on these parameters, only high-scored ASOs were included in the further analysis.
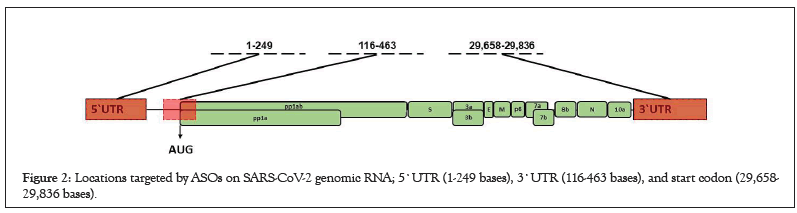
Figure 2: Locations targeted by ASOs on SARS-CoV-2 genomic RNA; 5`UTR (1-249 bases), 3`UTR (116-463 bases), and start codon (29,658- 29,836 bases).
ASO analysis
Off-target risks for high-scored ASOs were evaluated on the GenBank database by using the BLAST tool. ASO candidates with a high similarity rate (E-value<10) on the human genome database were eliminated.
Free energy values were used to calculate the binding energy of ASO-target complexes. In this context, free energy values for ASO-target complexes and free energy for SARS-CoV-2 genomic RNA were determined on RNA structure server (version 4.5). The binding energy of ASO-target complexes was identified using the equation below.
ASO candidates with a higher binding energy of ASO-target complexes than-20 kcal/mol were also eliminated since they will be probably inefficient as described by Aartsma-Rus et al. previously.
ASO modifications
Lastly, promising ASOs were then modified to improve their efficiency in the cell. All these modifications were determined and conducted based on literature (Figure 3).
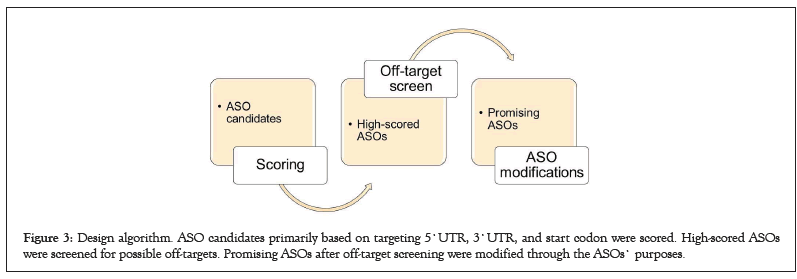
Figure 3: Design algorithm. ASO candidates primarily based on targeting 5`UTR, 3`UTR, and start codon were scored. High-scored ASOs were screened for possible off-targets. Promising ASOs after off-target screening were modified through the ASOs` purposes.
To design Antisense Oligonucleotides (ASOs) targeting SARSCoV- 2 genomic RNA, I obtained firstly reference sequence of SARS-CoV-2 genomic RNA (GenBank; MT385474). ASOs are chemically modified oligonucleotides with base-complementary abilities to a specific RNA target in the cell. ASOs designed near to 5`UTR, start codon, and or 3`UTR of the targeted gene can regulate the translation of genetic material into functional proteins. Particularly, the suppression of translation is achieved through a translational blockage or recruiting sequence-specific nucleases in a sequence-specific manner upon binding of ASOs to complementary RNA [22]. Thus, secondary structures of three viral regions on SARS-CoV-2 genomic RNA; 5`UTR, start codon, and or 3`UTR as the target in ASO design were predicted using mfold server to detect theoretical open regions available for ASO complementary. Two different conformations with the lowest free energy were taken as the template for each target site, and 24 ASO candidates targeting the open regions were designed in total (Tables S1 and S2).
I picked up the seven ASOs with a high potential to inhibit SARSCoV- 2 replication and transcription by inducing cleavage of the viral genomic RNA among 24 ASO candidates based on the parameters as described by Aartsma-Rus et al. previously (Figures 4a and 4b). All these seven ASOs have 20-24 nucleotides (nt) in length which is an optimum size for synthesis and long enough on statistical grounds to specify in the human genome (Table 1). Moreover, previous studies showed that the basic melting temperature (Tm) was significantly higher for the effective group of ASOs. Thereby, Tm values for ASOs were higher than 50°C in high-scored ASOs as well. Also, it is known that C and G content correlates with ASOtarget RNA duplex stability, and the total number of Gs and Cs was significantly higher in effective ASOs. Therefore, I kept C and G content more than 6 for each high-scored ASOs or more than 30% in particular. I also screened high-scored ASOs in the case of the ability to form stable secondary structures and dimerization since it has a negative effect on ASO efficacy and eliminated them. Because dimers and secondary structures will probably decrease ASO efficacy by blocking their complementary with the target (Figure 5) [23].
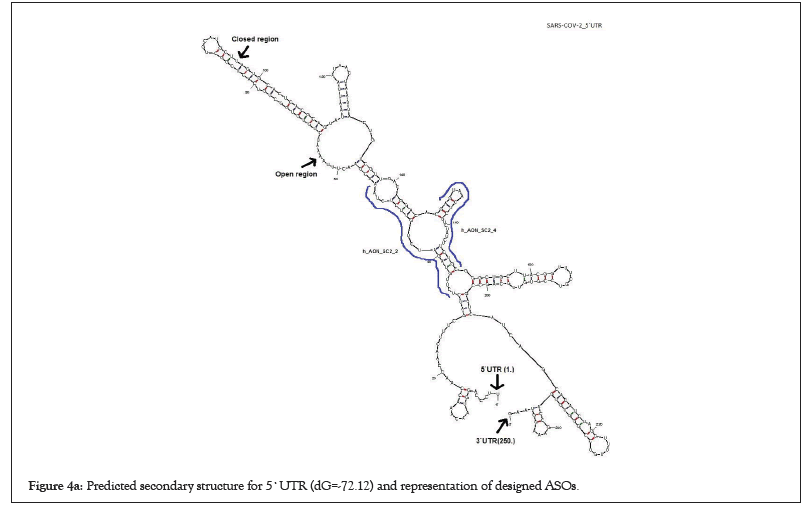
Figure 4a: Predicted secondary structure for 5`UTR (dG=-72.12) and representation of designed ASOs.
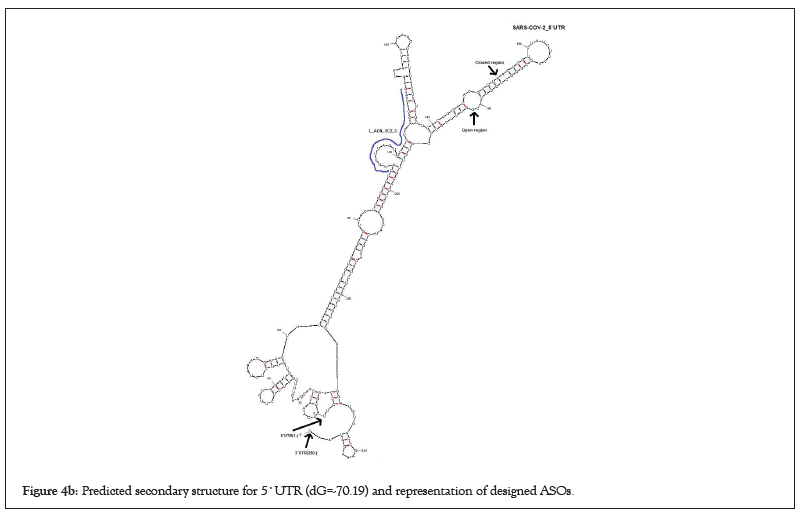
Figure 4b: Predicted secondary structure for 5`UTR (dG=-70.19) and representation of designed ASOs.
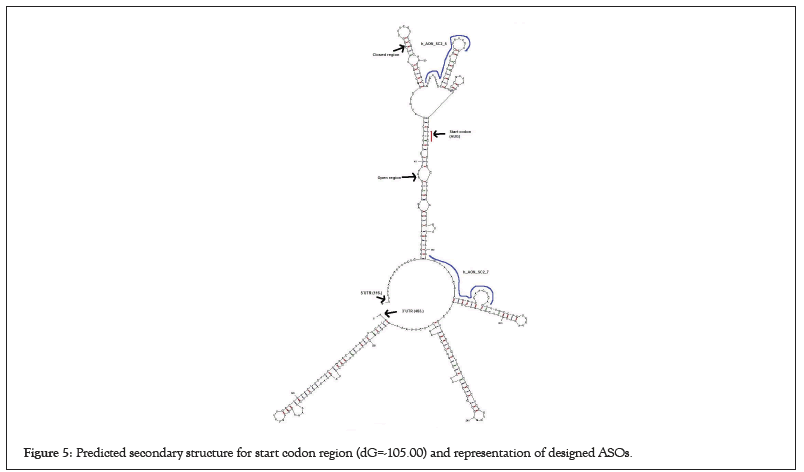
Figure 5: Predicted secondary structure for start codon region (dG=-105.00) and representation of designed ASOs.
| AONs | Name | Sense sequence (5`-->3`) |
Antisense sequence (5`-->3`) |
Target region | Length | Mw | Tm(°C) | GCs | GC% | Secondary structure | Dimer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| h_AON_SC2_2 | TR_1 | TCTTGTAGATCTGTTCTCTAA | UUAGAGAACAGAUCUACAAGA | 5`UTR | 21 | 6751,2 | 52,8 | 7 | 33,3 | Weak | No |
| h_AON_SC2_4 | TR_2 | AGTAACTCGTCTATCTTCTG | CAGAAGAUAGACGAGUUACU | 5`UTR | 20 | 6438 | 53,5 | 8 | 40 | None | No |
| h_AON_SC2_5 | TR_3 | ATCATCAGCACATCTAGGTTTTGT | ACAAAACCUAGAUGUGCUGAUGAU | Startcodon | 24 | 7684,7 | 59,6 | 9 | 37,5 | Very Weak | No |
| h_AON_SC2_7 | TR_4 | AGAAAACACACGTCCAACTCAG | CUGAGUUGGACGUGUGUUUUCU | Startcodon | 22 | 6990,1 | 59,4 | 10 | 45,5 | None | No |
| h_AON_SC2_11 | TR_5 | AGTGAACAATGCTAGGGAGA | UCUCCCUAGCAUUGUUCACU | 3`UTR | 20 | 6201,7 | 59,2 | 9 | 45 | None | No |
| L_AON_SC2_3 | TR_6 | ATGCTTAGTGCACTCACGCAG | CUGCGUGAGUGCACUAAGCAU | 5`UTR | 21 | 6712,1 | 63,1 | 11 | 52,4 | Weak | No |
| L_AON_SC2_9 | TR_7 | ATCGAGTGTACAGTGAAC | GUUCACUGUACACUCGAUCG | 3`UTR | 20 | 6303,8 | 57,1 | 10 | 50 | None | No |
Table 1: High-scored antisense oligonucleotides based on the parameters described previously.
Off-target risks on the human transcriptome were checked through the BLAST tool. The ASOs with high similarity with any gene on the human transcriptome were left out of the list to avoid possible side effects due to the antisense therapy. Then, high-scored ASOs with a low off-target risks were handled for further modifications.
Natural phosphodiester oligonucleotides are quickly digested by cellular nucleases in the cell [24]. Therefore, ASOs were modified to some forms that are nuclease-resistant and appropriate for research purposes (Table 2). Several modifications such as 5-methyl deoxycytosine (5Me-dC) deaza G (7-deaza-dG) 2'-O-methoxy-ethyl 5me Uridine (2'-O-MOE-RNA) Phosphorothioate linkage (PS) have been applied for high-scored ASOs to modify them through a research purpose and more resistant to cellular nucleases (Figures 6a and 6b) [12,25].
| Name | Modified structures |
|---|---|
| TR_1 | U*U*A*G*A*G*A*A*C*A*G*A*U*C*U*A*C*A*A*G*A |
| TR_2 | C*A*G*A*A*G*A*U*A*G*A*mC*G*A*G*U*U*A*C*U |
| TR_3 | A*C*A*A*A*A*xC*xC*U*A*G*A*U*G*U*G*C*U*G*A*U*G*A*U |
| TR_4 | C*U*G*A*G*U*U*xG*xG*A*mC*G*U*G*U*G*U*U*U*U*C*U |
| TR_5 | U*C*U*xC*xC*xC*U*A*G*C*A*U*U*G*U*U*C*A*C*U |
| TR_6 | C*U*G*mC*G*U*G*A*G*U*G*C*A*C*U*A*A*G*C*A*U |
| TR_7 | G*U*U*C*A*C*U*G*U*A*C*A*C*U*mC*G*A*U*mC*G |
Note: Underline=2'-O-MOE-RNA (MOE is 2-methoxyethyl), m=5-Me-dC, *=PS, x=7-deaza-dG.
Table 2: Antisense oligonucleotides with high-score in modified forms.
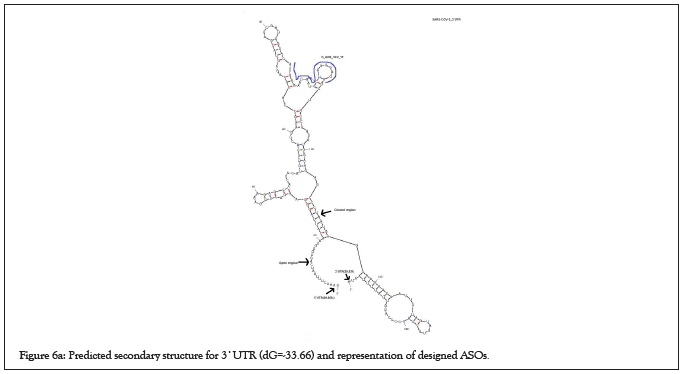
Figure 6a: Predicted secondary structure for 3`UTR (dG=-33.66) and representation of designed ASOs.
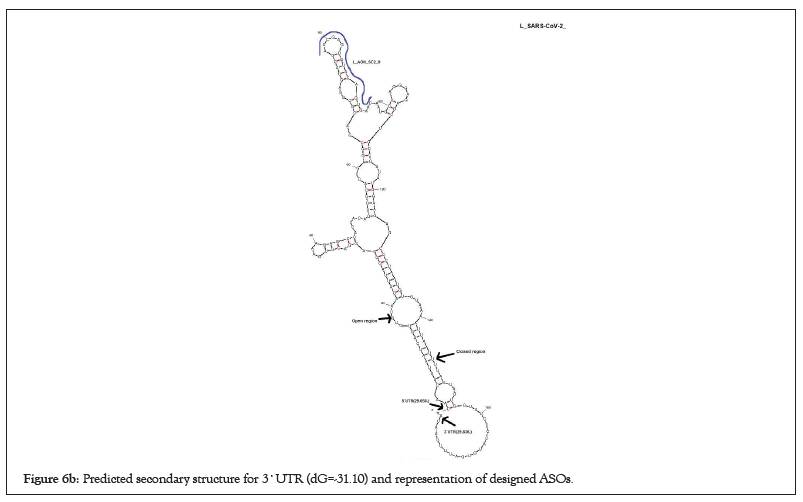
Figure 6b: Predicted secondary structure for 3`UTR (dG=-31.10) and representation of designed ASOs.
PS that link nucleotides through a phosphate backbone variant facilitates ASO entry into the cell since they retain negative backbone charges. Moreover, PS linkages are nuclease resistant, providing longer plasma half-lives in the cell. On the contrary, they have several negative impacts on ASO activity such as reduced hybridization to target mRNAs, phosphorothioate interaction with proteins, therefore lead to negative side effects, including immune system activity. For this purpose, all ASOs are modified with PS internucleotide linkages to protect them from cellular nucleases (Figure 7a) [26].
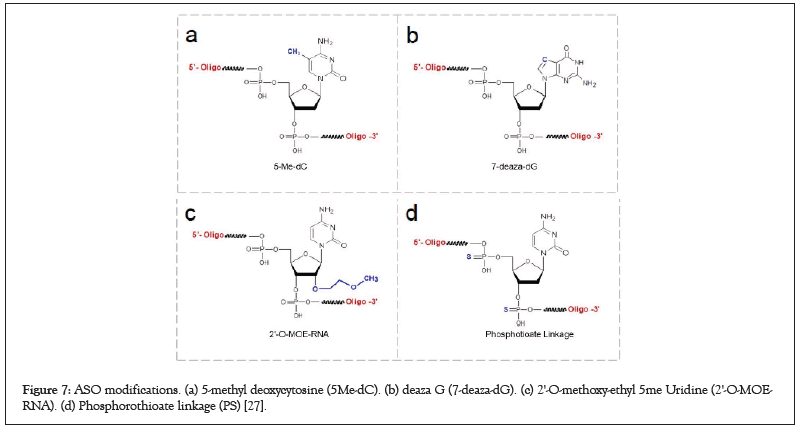
Figure 7: ASO modifications. (a) 5-methyl deoxycytosine (5Me-dC). (b) deaza G (7-deaza-dG). (c) 2'-O-methoxy-ethyl 5me Uridine (2'-O-MOERNA). (d) Phosphorothioate linkage (PS) [27].
2'-O-MOE-RNA (MOE is 2-MethOxyEthyl) is a type of enhancedbinding- affinity sugar modification. ASOs containing 2′-MOE modifications have enhanced nuclease resistance with lower cell toxicity and increased binding affinity to the target sequences. However, ASOs fully including sugar-modified RNA-like nucleotides (such as 2′-MOE) do not support RNase H cleavage of the complementary RNA, therefore ASOs should be constructed in the hetero form to induce RNase H cleavage. In this case, TR_1, 3, and 7 were modified by 2′-MOE modifications to increase binding affinity and decrease cell toxicity (Figure 7c) [26].
Also, ASOs involving unmethylated CpG islands may stimulate the immune system like that of bacterial DNA as the CpG motifs in the bacterial genome triggers the activation of immune cells. To get rid of immunostimulation, designed ASOs should not include a high amount of CpG islands or if it involves, it must be modified the cytosines in CpG with 5’-methylcytosine, which has been shown to decrease immunostimulation significantly. TR_2, 4, 6 which have at least one CpG island were modified by replacing cytosines with 5’-methylcytosine (Figure 7d) [26].
Lastly, ASOs containing two or more C or G nucleotides can form inappropriate structures, which might trigger undesirable, off-target effects. Previous studies have shown that polyG stretches of G bases are able to lead the formation of G-quartets, which may help binding to proteins, by the way, transcription factors, thereby interfere with antisense activity. To avoid G-quartets, 7-deaza-dG modifications to replace G bases which will probably prevent quartet formation. PolyG including TR_3, 4, and 5 ASOs undergone modifications to replace G bases to 7-deaza-dG modified forms (Figure 7b) [26].
In the final stage of the study, I evaluated the binding energy of ASO-target complexes to identify their efficiency in silico [13]. To do this, free energy values were used to calculate the binding energy of AON-target complexes as described in the methods. Aartsma- Rus at el. previously demonstrated that the binding energy of ASOtarget complexes was significantly higher for effective ASOs (< -20, respectively). In this line, our results showed that all ASOs have less binding free energy than-20 kcal/mol (Figure 8) [27,28].
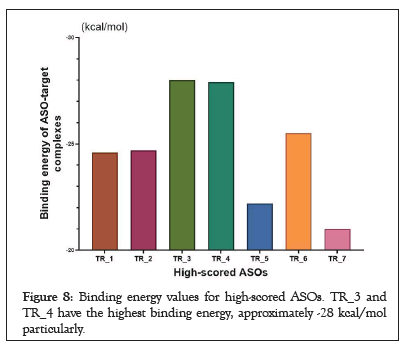
Figure 8: Binding energy values for high-scored ASOs. TR_3 and TR_4 have the highest binding energy, approximately -28 kcal/mol particularly.
Overall, this study hidhlighted that antisense theraphy could be a novel strategy to threat present COVID-19 pandemic by targeting SARS-CoV-2 genomic RNA directly. In this context, ASOs targeting 5`UTR, start codon, and or 3`UTR of the targeted gene were designed and eliminated low-scored ASO candidates. Seven highscored ASOs based on parameters described in the literature were determined as a novel for future studies. Those seven promising ASOs can be synthesized followed by required modifications and test on SARS-CoV-2 infected Vero cells to screen their efficacy for the inhibition of viral replication and transcription.
I would like to thank Ozlem Yalcin Capan, Asst. Prof. for assistance with her guide during paper publishing, and for her comments that greatly improved the manuscript. I would also like to show my gratitude to the Nazlican Yurekli, BSc for sharing her pearls of wisdom with me during the course of this research. I am also immensely grateful to my family for their support.
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
[Cross ref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Cross ref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Cross ref] [Google scholar] [PubMed]
[Cross ref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Cross ref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Cross ref] [Google scholar] [PubMed]
[Cross ref] [Google scholar] [PubMed]
[Cross ref] [Google scholar] [PubMed]
[Cross ref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
Citation: Cubuk H (2022) A Promising Strategy against SARS-CoV-2 Infected Patients: Antisense Therapy. Transcriptomics-Open Access. 8:119.
Received: 30-Jun-2022, Manuscript No. TOA-22-18170; Editor assigned: 04-Jul-2022, Pre QC No. TOA-22-18170 (PQ); Reviewed: 18-Jul-2022, QC No. TOA-22-18170; Revised: 25-Jul-2022, Manuscript No. TOA-22-18170 (R); Published: 01-Aug-2022 , DOI: 10.35248/2329-8936.22.8.119
Copyright: © 2022 Cubuk H. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.