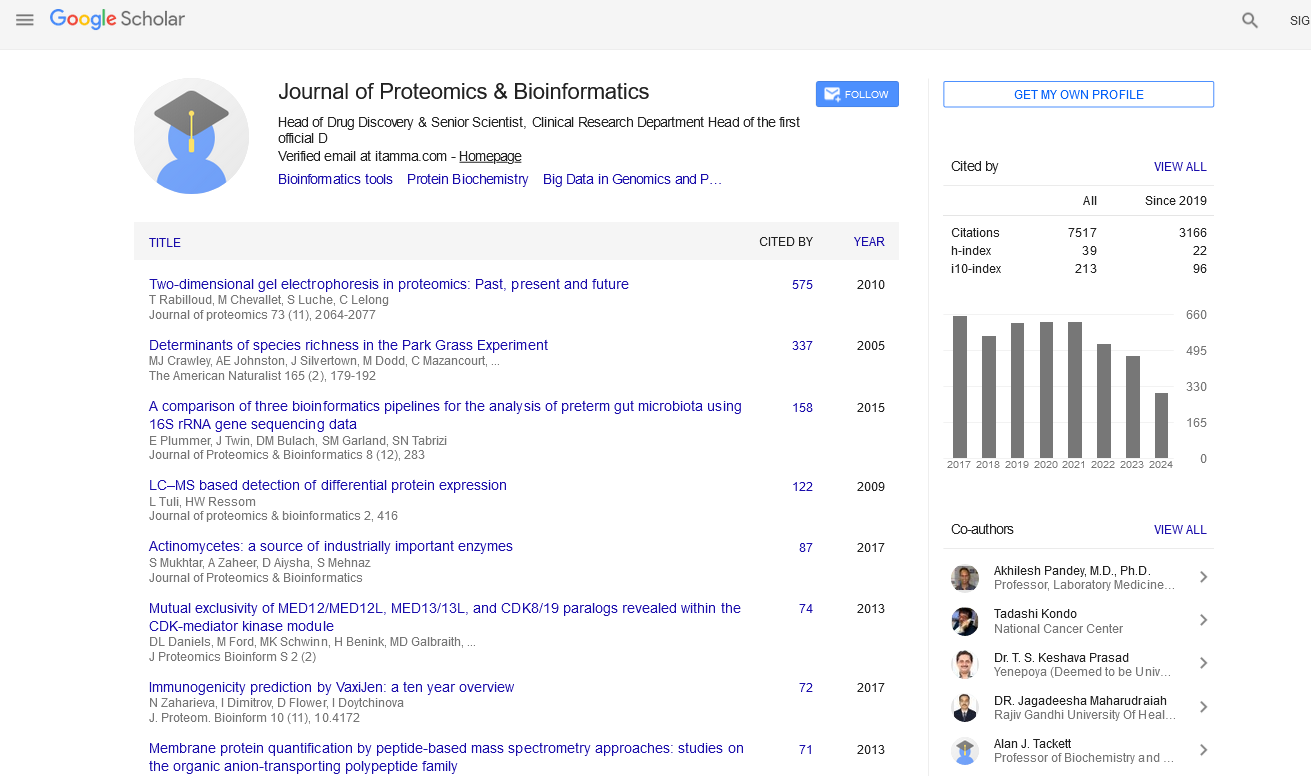
Journal of Proteomics & Bioinformatics : Citations & Metrics Report
Articles published in Journal of Proteomics & Bioinformatics have been cited by esteemed scholars and scientists all around the world. Journal of Proteomics & Bioinformatics has got h-index 39, which means every article in Journal of Proteomics & Bioinformatics has got 39 average citations.
Following are the list of articles that have cited the articles published in Journal of Proteomics & Bioinformatics.
| 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | |
|---|---|---|---|---|---|---|---|---|---|
Total published articles |
23 | 37 | 61 | 64 | 15 | 21 | 31 | 45 | 47 |
Research, Review articles and Editorials |
9 | 17 | 11 | 37 | 11 | 12 | 28 | 40 | 37 |
Research communications, Review communications, Editorial communications, Case reports and Commentary |
1 | 17 | 50 | 25 | 3 | 9 | 3 | 2 | 4 |
Conference proceedings |
0 | 0 | 15 | 0 | 0 | 0 | 161 | 354 | 225 |
Citations received as per Google Scholar, other indexing platforms and portals |
0 | 0 | 0 | 524 | 565 | 544 | 497 | 546 | 567 |
| Journal total citations count | 7517 |
| Journal impact factor | 10.40 |
| Journal 5 years impact factor | 16.58 |
| Journal cite score | 18.65 |
| Journal h-index | 39 |
| Journal h-index since 2019 | 22 |
Important citations (453)
ceciliani f, roccabianca p, giudice c, lecchi c (2016) application of post-genomic techniques in dog cancer research. mol biosyst 12:2665-2679. |
|
maria-neto s, de almeida kc, macedo ml, franco ol (2015) understanding bacterial resistance to antimicrobial peptides: from the surface to deep inside. biochimica et biophysica acta. 1848:3078-3088. |
|
xiao x, wu yt (2015) irspot-gcfpsenc: identifying recombination spots with pseudo nucleotide composition. biomedical engineering and environmental engineering 6:3. |
|
tiwari ak, srivastava r (2018) an efficient approach for prediction of nuclear receptor and their subfamilies based on fuzzy k-nearest neighbor with maximum relevance minimum redundancy. proc natl acad sci india a 88:129-136. |
|
tiwari ak, srivastava r (2015) feature based classification of nuclear receptors and their subfamilies using fuzzy k-nearest neighbor. in2015 international conference on advances in computer engineering and applications. ieee pp. 24-28. |
|
zhang l, zhang c, gao r, yang r (2015) jppred: prediction of types of j-proteins from imbalanced data using an ensemble learning method. biomed res int 2015. |
|
ismail hd, saigo h, kc db (2018) rf-nr: random forest based approach for improved classification of nuclear receptors. ieee/acm transactions on computational biology and bioinformatics (tcbb) 15:1844-1852. |
|
zhang l, zhang c, gao r, yang r (2015) incorporating g-gap dipeptide composition and position specific scoring matrix for identifying antioxidant proteins. in2015 ieee 28th canadian conference on electrical and computer engineering (ccece). ieee pp. 31-36. |
|
nebel jc (2014) computational intelligence in bioinformatics. j proteomics bioinform s 9:2. |
|
tiwari ak, srivastava r (2015) an efficient approach for the prediction of ion channels and their subfamilies. comput biol chem 58:205-221. |
|
zhang l, zhang c, gao r, yang r, song q (2016) sequence based prediction of antioxidant proteins using a classifier selection strategy. plos one 11:e0163274. |
|
zhang l, zhang c, gao r, yang r, song q (2016) using the smote technique and hybrid features to predict the types of ion channel-targeted conotoxins. j theor biol 403:75-84. |
|
zhang l, zhang c, gao r, yang r (2015) an ensemble method to distinguish bacteriophage virion from non-virion proteins based on protein sequence characteristics. int j mol sci 16:21734-21758. |
|
tiwari ak, srivastava r (2014) a survey of computational intelligence techniques in protein function prediction. international journal of proteomics 2014. |
|
ng s (2019) multi-omics analysis of immune-metabolic blood responses of very preterm infants for improving diagnosis of late-onset sepsis (doctoral dissertation, murdoch university). |
|
rusconi b, good m, warner bb (2017) the microbiome and biomarkers for necrotizing enterocolitis: are we any closer to prediction? j pediatr 189:40-47. |
|
samuel bo, folashade o (2018) nanoscale science and nanotechnology education in africa: importance and challenges. afr j chem educ 8:7-27. |
|
ng s, strunk t, jiang p, muk t, sangild pt, currie a, et al. (2018) precision medicine for neonatal sepsis. front mol biosci 5:70. |
|
rusconi b, good m, warner bb (2017) the microbiome and biomarkers for necrotizing enterocolitis: are we any closer to prediction? j pediatr 189:40-47. |
|
riveros c, ujaldon m, moscato p (2016) gpu acceleration of an entropy-based model to quantify epistatic interactions between snps. curr bioinform 11:396-407. |
|