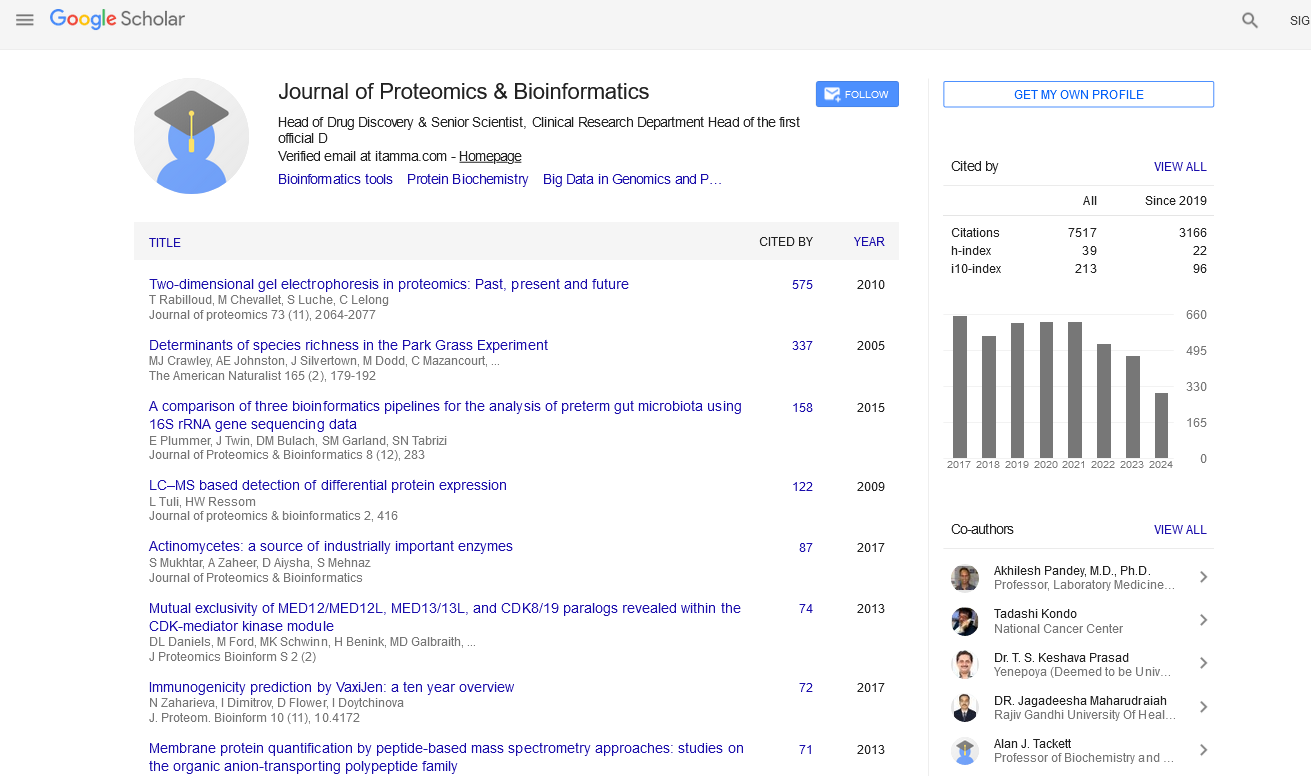
Journal of Proteomics & Bioinformatics : Citations & Metrics Report
Articles published in Journal of Proteomics & Bioinformatics have been cited by esteemed scholars and scientists all around the world. Journal of Proteomics & Bioinformatics has got h-index 39, which means every article in Journal of Proteomics & Bioinformatics has got 39 average citations.
Following are the list of articles that have cited the articles published in Journal of Proteomics & Bioinformatics.
| 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | |
|---|---|---|---|---|---|---|---|---|---|
Total published articles |
23 | 37 | 61 | 64 | 15 | 21 | 31 | 45 | 47 |
Research, Review articles and Editorials |
9 | 17 | 11 | 37 | 11 | 12 | 28 | 40 | 37 |
Research communications, Review communications, Editorial communications, Case reports and Commentary |
1 | 17 | 50 | 25 | 3 | 9 | 3 | 2 | 4 |
Conference proceedings |
0 | 0 | 15 | 0 | 0 | 0 | 161 | 354 | 225 |
Citations received as per Google Scholar, other indexing platforms and portals |
0 | 0 | 0 | 524 | 565 | 544 | 497 | 546 | 567 |
| Journal total citations count | 7517 |
| Journal impact factor | 10.40 |
| Journal 5 years impact factor | 16.58 |
| Journal cite score | 18.65 |
| Journal h-index | 39 |
| Journal h-index since 2019 | 22 |
Important citations (453)
Mahmud m, kaiser ms, hussain a, vassanelli s (2018) applications of deep learning and reinforcement learning to biological data. ‎ieee trans neural netw learn syst 29:2063-2079. |
|
Meech r, hu dg, mckinnon ra, mubarokah sn, haines az, nair pc, et al. (2019) superfamily: new members, new functions, and novel paradigms. physiol rev 99:1153–1222. |
|
Krishna gv (2017) department of biotechnology. j food sci res 2:110. |
|
Holmes rs (2017) vertebrate arylsulfatase k (arsk): comparative and evolutionary studies of the lysosomal 2-sulfoglucuronate sulfatase. j data mining genomics proteomics 8:2153-0602. |
|
Holmes rs (2017) comparative and evolutionary studies of vertebrate extracellular sulfatase genes and proteins: sulf1 and sulf2. j proteomics bioinform 10:32-40. |
|
Holmes rs (2017) comparative studies of vertebrate iduronate 2-sulfatase (ids) genes and proteins: evolution of a mammalian x-linked gene. 3 biotech 7:22. |
|
Meech r, hu dg, mckinnon ra, mubarokah sn, haines az, nair pc, et al. (2019) the udp-glycosyltransferase (ugt) superfamily: new members, new functions, and novel paradigms. physiol rev 99:1153-1222. |
|
krishna gv (2017) department of biotechnology. j food sci res 2:110. |
|
Ghosh r, upadhayay ad, roy ak (2017) in silico analysis, structure modeling and phosphorylation site prediction of vitellogenin protein from gibelion catla. jappl biotechnol bioeng 3:00055. |
|
Ghosh r, upadhayay ad, roy ak (2017) in silico analysis, structure modeling and phosphorylation site prediction of vitellogenin protein from gibelion catla. jappl biotechnol bioeng 3:00055. |
|
rumpi g, upadhyay ad, roy ak, samik a (2017) structural analysis of cytochrome c genes of major carp and utility of forensic investigation. j forensic crime investi 1:103. |
|
Dutta b, banerjee a, chakraborty p, bandopadhyay r (2018) in silico studies on bacterial xylanase enzyme: structural and functional insight. genet eng biotechnol j 16:749-756. |
|
krishna gv (2017) department of biotechnology. j food sci res 2:110. |
|
Лашков ÐÐ, РубинÑкий СВ, ÐйÑтрих-Геллер ПР(2019) Применение алгоритма dbscan Ð´Ð»Ñ Ð²Ñ‹ÑÐ²Ð»ÐµÐ½Ð¸Ñ Ð³Ð¸Ð´Ñ€Ð¾Ñ„Ð¾Ð±Ð½Ñ‹Ñ… клаÑтеров в Ñтруктурах белков. КриÑÑ‚Ð°Ð»Ð»Ð¾Ð³Ñ€Ð°Ñ„Ð¸Ñ 64:494-502. |
|
Lashkov aa, rubinsky sv, eistrikh-heller pa (2019) application of the dbscan algorithm to detect hydrophobic clusters in protein structures. crystallogr rep 64:524-532. |
|
krishna gv (2017) department of biotechnology. j food sci res 2:110. |
|
banach m, kalinowska b, konieczny l, roterman i (2018) possible mechanism of amyloidogenesis of v domains. self-assembled molecules—new kind of protein ligands—supramolecular ligands; roterman i, konieczny l, eds. 2018:77-100. |
|
krishna gv (2017) department of biotechnology. j food sci res 2:110. |
|
krishna gv (2017) department of biotechnology. j food sci res 2:110. |
|
jariyal h, gupta c, bhat vs, wagh jr, srivastava a (2019) advancements in cancer stem cell isolation and characterization. stem cell rev rep 2019:1-9. |
|