Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2008) Volume 1, Issue 4
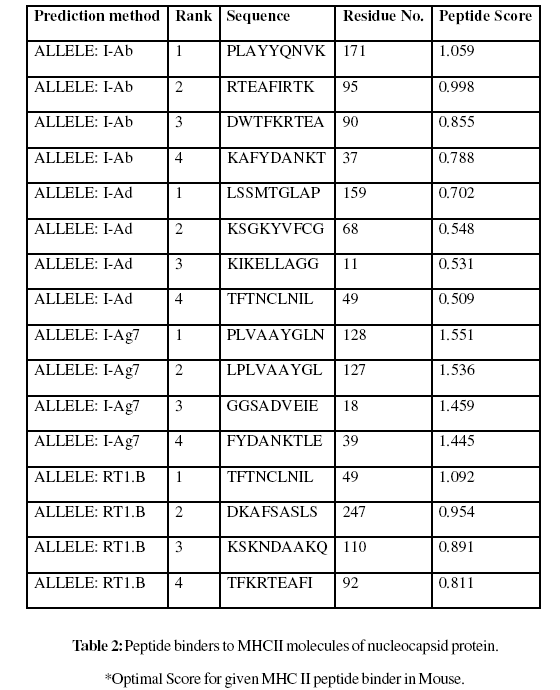
A virus disease caused of crop losses to groundnut in Andhra Pradesh during kharif in the year 2000. It was presumed to be caused by Groundnut bud necrosis virus (GBNV), a virus known to be widely distributed in India. It is one among the important infestations affecting groundnut cultivation especially during summer. Nucleocapsid peptides of groundnut bud necrosis virus are most suitable for subunit vaccine development because with single epitope, the immune response can be generated in large population. Analysis shows MHC class II binding peptides of coat protein from SMV are important determinant for protection of many plants form viral infection. In this assay we predicted the binding affinity of SMV genome polyprotein having 276 amino acids, which shows 268 nonamers. In this analysis, we found the SVM based MHCII-IAb peptide regions, 171- PLAYYQNVK, 95- RTEAFIRTK, 90- DWTFKRTEA, 37- KAFYDANKT, (optimal score is 1.059); MHCII-IAd peptide regions, 159- LSSMTGLAP, 68- KSGKYVFCG, 11- KIKELLAGG, 49- TFTNCLNIL, (optimal score is 0.702); MHCIIIAg7 peptide regions, 128- PLVAAYGLN, 127- LPLVAAYGL, 18- GGSADVEIE, 39- FYDANKTLE, (optimal score is 1.551); and MHCII- RT1.B peptide regions, 49- TFTNCLNIL, 247- DKAFSASLS, 110- KSKNDAAKQ, 92- TFKRTEAFI , (optimal score is 1.092) which represented predicted binders from nucleocapsid protein. The method integrates prediction of peptide MHC class I binding; proteasomal C terminal cleavage and TAP transport efficiency of the nucleocapsid protein of GBNV. Thus a small fragment of antigen can induce immune response against whole antigen. This theme is implemented in designing subunit and synthetic peptide vaccines.
Keywords: Groundnut Bud Necrosis Virus; Antigenic Peptides; MHC-Binders; SVM; Nonamers
This is a virus disease causing more damage to groundnut crop during kharif and summer. Virus is mainly transmitted through thrips (Thrips palmae). Infected plants appear bushy and stunted during early stages Groundnut Bud Necrosis (GBN) is the most threatening viral disease of groundnut. It is one among the important infestations affecting groundnut cultivation especially during summer. The disease is caused by the Groundnut bud necrosis virus (GBNV). During initial stages of infection mild spots are formed, which later develop into chlorotic rings. Necrosis of the terminal bud, a characteristic symptom occurs on crop grown during the monsoon and shortly after. Terminal bud Necrosis occurs when temperature is relatively high. As plant matures, it becomes stunted with short internodes and proliferation of Axillary shoots. Petioles bearing fully expanded leaflets with initial symptoms become flaccid and droop. This symptoms is followed by terminal bud necrosis. Primary symptoms are followed by secondary symptoms. These are stunting and proliferation of axillary shoots. Leaflets formed on the axillary shoots show a wide range of symptoms including reduction in size, distortion of the lamina, mosaic mottling and general chlorosis. These secondary symptoms are most common on early infected plants giving them a stunted and bushy appearance.
Virus Transmission
The virus is transmitted by thrips, which have a wide range of hosts. The virus survives in these hosts and acts as a source of inoculums for the vector. The thrips are carried by wind. The population of vectors increases rapidly from January-March and August-September Kharif and hence the crop suffers a heavy loss in both the seasons. A prolonged dry spell favours the multiplication of thrips and spread of the virus.
Strategy
The phenotype of the resistant transgenic plants includes fewer centers of initial virus infection, a delay in symptom development, and low virus accumulation. Protoplasts from virus resistant transgenic plants are also resistant, suggesting that the protection is largely operational at the cellular level. Transgenic plants expressing nucleocapsid protein are protected against infection by virus particles but are susceptible to viral RNA, indicating that the protection may primarily involve an inhibition of virus uncoating. This approach is based on the phenomenon of cross-protection (Valkonen et al., 2002) hereby a plant infected with a mild strain of virus is protected against a more severe strain of the same virus. Proteins of soybean mosaic virus are necessary for its production in or on all food commodities. An exemption from the requirement of a tolerance is established for residues of the biological plant pesticide.
MHC Class Binding Peptides
The new paradigm in vaccine design is emerging, following essential discoveries in immunology and development of new MHC Class-I binding peptides prediction tools (Bhasin et al., 2003; Singh and Raghava, 2002; Cui et al., 2006; Julia and Philip, 2007). MHC molecules are cell surface glycoproteins, which take active part in host immune reactions. The involvement of MHC class-I in response to almost all antigens and the variable length of interacting peptides make the study of MHC Class I molecules very interesting. MHC molecules have been well characterized in terms of their role in immune reactions. They bind to some of the peptide fragments generated after proteolytic cleavage of antigen (Kumar et al., 2007). This binding acts like red flags for antigen specific and to generate immune response against the parent antigen. So a small fragment of antigen can induce immune response against whole antigen. Nucleocapsid peptides are most suitable for subunit vaccine development because with single epitope, the immune response can be generated in large population. MHCpeptide complexes will be translocated on the surface of antigen presenting cells (APCs). This theme is implemented in designing subunit and synthetic peptide vaccines (Gomase et al., 2007). One of the important problems in subunit vaccine design is to search antigenic regions in an antigen (Schirle et al., 2001) that can stimulate T cells called T-cell epitopes. In literature, fortunately, a large amount of data about such peptides is available. Pastly and presently, a number of databases have been developed to provide comprehensive information related to T-cell epitopes (Rammensee et al., 1999; Blythe et al., 2002; Schonbach et al., 2002 and Korber et al., 2001)
Protein Sequence Analysis
The nucleocapsid protein sequence of Groundnut bud necrosis virus was analyzed to study the antigenicity (Saritha and Jain, 2007), solvent accessible regions and MHC class peptide binding, which allows potential drug targets to identify active sites against plant diseases.
Prediction of Antigenicity
Prediction of antigenicity program predicts those segments from within viral pathogenicity protein that are likely to be antigenic by eliciting an antibody response. Antigenic epitopes is determined using the Gomase , (2007), Hopp and Woods, Welling, Parker, B-EpiPred Server and Kolaskar and Tongaonkar antigenicity methods (Gomase, 2006; Hopp and Woods,1981; Welling et al., 1985; Jens Erik et al., 2006; Parker et al., 1986 and Kolaskar and Tongaonkar, 1990)
Prediction of Protein Secondary Structure
The important concepts in secondary structure prediction are identified as: residue conformational propensities, sequence edge effects, moments of hydrophobicity, position of insertions and Deletions in aligned homologous sequence, moments of conservation, auto-correlation, residue ratios, secondary structure feedback effects, and filtering (Garnier, 1978; and Robson and Garnier, 1993).
Finding the Location in Solvent Accessible Regions
Finding the location in solvent accessible regions in protein, type of plot determines the hydrophobic and hydrophilic scales and it is utilized for prediction. This may be useful in predicting membrane-spanning domains, potential antigenic sites and regions that are likely exposed on the protein surface (Sweet and Eisenberg, 1983; Kyte and Doolittle, 1982; Abraham and Leo, 1987;Bull and Breese, 1974; Guy, 1985; Miyazawa and Jernigen, 1985; Roseman, 1988; Wolfenden et al., 1981; Wilson et al., 1981; Aboderin, 1971; Chothia, 1976; Eisenberg et al., 1984; Manavalan and Ponnuswamy, 1978;Black and Mould, 1991; Fauchere and Pliska, 1983; Janin, 1979; Rao and Argos, 1986; Tanford, 1962;Cowan and Whittaker, 1990; Rose et al., 1985; Wilkins et al., 1999 and Eisenberg et al., 1984)
Prediction of MHC Binding Peptide
The MHC peptide binding is predicted using neural networks trained on C terminals of known epitopes. In analysis predicted MHC/peptide binding is a log-transformed value related to the IC50 values in nM units. MHC2Pred predicts peptide binders to MHCI and MHCII molecules from protein sequences or sequence alignments using Position Specific Scoring Matrices (PSSMs). Support Vector Machine (SVM) based method for prediction of promiscuous MHC class II binding peptides. The average accuracy of SVM based method for 42 alleles is ~80%. For development of MHC binder, an elegant machine learning technique SVM has been used. SVM has been trained on the binary input of single amino acid sequence. In addition, we predicts those MHCI ligands whose C-terminal end is likely to be the result of proteosomal cleavage (Brusic et al., 1998; Bhasin and Raghava, 2005Gomase et al., 2008).
A nucleocapsid sequence is 276 residues long as
MSNVKQLTEKKIKELLAGGSADVEIETEDSTPGFSFKAFYDANKTLEITFTNCLNILKCRK QIFAACKSGKYVFCGKTIVATNTDVGPDDWTFKRTEAFIRTKMASMVEKSKNDAAKQE MYNKIMELPLVAAYGLNVPASFDTCALRMMLCIGGPLPLLSSMTGLAPIIFPLAYYQNV KKEKLGIKNFSTYEQVCKVAKVLSASQIEFKNELEVMFKSAVKLLSESNPGTASSISLKK YDEQVKYMDKAFSASLSMDDYGEHSKKKSSKAGPSLEL
Prediction of Antigenic Peptides
In these methods we found the antigenic determinants by finding the area of greatest local hydrophilicity. The HoppWoods scale was designed to predict the locations of antigenic determinants in a protein, assuming that the antigenic determinants would be exposed on the surface of the protein and thus would be located in hydrophilic regions (Fig 2). Its values are derived from the transfer-free energies for amino acid side chains between ethanol and water. Welling antigenicity plot gives value as the log of the quotient between percentage in a sample of known antigenic regions and percentage in average proteins (Fig 3). We also study B-EpiPred Server, Parker, Kolaskar and Tongaonkar antigenicity methods and the predicted antigenic fragments can bind to MHC molecule is the first bottlenecks in vaccine design (Fig 4-6).
The Robson and Garnier method predicted the secondary structure of the nucleocapsid protein. Each residue is assigned values for alpha helix, beta sheet, turns and coils using a window of 7 residues (fig 7). Using these information parameters, the likelihood of a given residue assuming each of the four possible conformations alpha, beta, reverse turn, or coils calculated, and the conformation with the largest likelihood is assigned to the residue.
Solvent Accessible Regions
Solvent accessible scales for delineating hydrophobic and hydrophilic characteristics of amino acids and scales are developed for predicting potential antigenic sites of globular proteins, which are likely to be rich in charged and polar residues. It was shown that a nucleocapsid protein is hydrophobic in nature and contains segments of low complexity and high-predicted flexibility (fig: - 8-28).
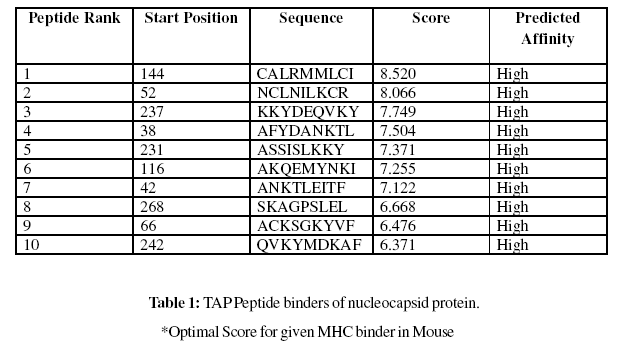
Prediction of MHC Binding Peptides
These MHC binding peptides are sufficient for eliciting the desired immune response. The prediction is based on cascade support vector machine, using sequence and properties of the amino acids. The correlation coefficient of 0.88 was obtained by using jack-knife validation test. In this test, we found the MHCI and MHCII binding regions (Table-1,Table-2). MHC molecules are cell surface glycoproteins, which take active part in host immune reactions and involvement of MHC class-I and MHC II in response to almost all antigens. In this assay we predicted the binding affinity of nucleocapsid protein having 276 amino acids, which shows different nonamers (Table-1,Table-2). For development of MHC binder prediction method, an elegant machine learning technique support vector machine (SVM) has been used. SVM has been trained on the binary input of single amino acid sequence. In this assay we predicted the binding affinity of nucleocapsid protein sequence (IsTX) having 276 amino acids, which shows 268 nonamers. Small peptide regions found as 144- CALRMMLCI (score 8.520), 52- NCLNILKCR (Score- 8.066), 237- KKYDEQVKY (Score- 7.749), 38- AFYDANKTL (Score- 7.504), known as nucleocapsid protein TAP transporter (Table-1). We also found the SVM based MHCII-IAb peptide regions, 171- PLAYYQNVK, 95- RTEAFIRTK, 90- DWTFKRTEA, 37- KAFYDANKT, (optimal score is 1.059); MHCII-IAd peptide regions, 159- LSSMTGLAP, 68- KSGKYVFCG, 11- KIKELLAGG, 49- TFTNCLNIL, (optimal score is 0.702); MHCII-IAg7 peptide regions , 128- PLVAAYGLN, 127-LPLVAAYGL, 18- GGSADVEIE, 39- FYDANKTLE, (optimal score is 1.551); and MHCII- RT1.B peptide regions, 49- TFTNCLNIL, 247- DKAFSASLS, 110- KSKNDAAKQ, 92- TFKRTEAFI , (optimal score is 1.092) which represented predicted binders from nucleocapsid protein. (Table-2). The predicted binding affinity is normalized by the 1% fractil. The MHC peptide binding is predicted using neural networks trained on C terminals of known epitopes. In analysis predicted MHC/peptide binding is a log-transformed value related to the IC50 values in nM units. These MHC binding peptides are sufficient for eliciting the desired immune response. Predicted MHC binding regions in an antigen sequence and there are directly associated with immune reactions, in analysis we found the MHCI and MHCII binding regions.
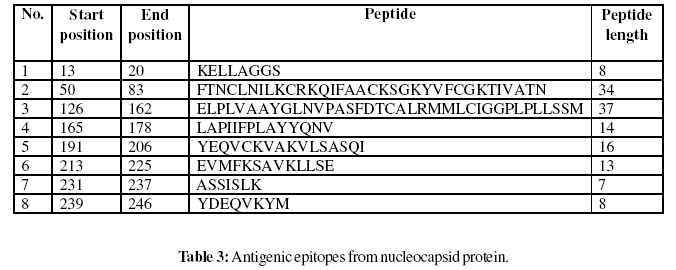
Gomase method (2007), B-EpiPred Server, Hopp and Woods, Welling, Parker, Kolaskar and Tongaonkar antigenicity scales were designed to predict the locations of antigenic determinants in Groundnut bud necrosis virus (nucleocapsid). Nucleocapsid shows beta sheets regions, which are high antigenic response than helical region of this peptide and shows highly antigenicity (Figure 1-5). We also found the Sweet hydrophobicity, Kyte & Doolittle hydrophobicity, Abraham & Leo , Bull & Breese hydrophobicity, Guy, Miyazawa hydrophobicity, Roseman hydrophobicity, Cowan HPLC pH7.5 hydrophobicity, Rose hydrophobicity, Eisenberg hydrophobicity, Manavalan hydrophobicity, Black hydrophobicity, Fauchere hydrophobicity, Janin hydrophobicity, Rao & Argos hydrophobicity, Wolfenden hydrophobicity, Wilson HPLC hydrophobicity, Cowan HPLC pH3.4, Tanford hydrophobicity, Rf mobility hydrophobicity and Chothia hydrophobicity scales, Theses scales are essentially a hydrophilic index, with a polar residues assigned negative values (Figures-7-27). In this assay we predicted the binding affinity of nucleocapsid protein having 41 amino acids, which shows 34 nonamers. Small peptide regions found as 144- CALRMMLCI (score 8.520), 52- NCLNILKCR (Score- 8.066), 237- KKYDEQVKY (Score- 7.749), 38- AFYDANKTL (Score- 7.504), known as nucleocapsid protein TAP transporter. Adducts of MHC and peptide complexes are the ligands for T cell receptors (TCR) (Table-1). MHC molecules are cell surface glycoproteins, which take active part in host immune reactions and involvement of MHC class-I and MHC II in response to almost all antigens (Table- 2). Kolaskar and Tongaonkar antigenicity are the sites of molecules that are recognized by antibodies of the immune system for the nucleocapsid protein, analysis shows epitopes present in the nucleocapsid protein the desired immune response (Table-3). The region of maximal hydrophilicity is likely to be an antigenic site, having hydrophobic characteristics, because C- terminal regions of nucleocapsid protein is solvent accessible and unstructured, antibodies against those regions are also likely to recognize the native protein.
For the prediction of antigenic determinant site of nucleocapsid protein, we got eighteen antigenic determinant sites in the sequence. The highest pick is recorded between sequence of amino acid in the region are ‘126- ELPLVAAYGLNVPASFDTCALRMMLCIGGPLPLLSSM- 162 and 50- FTNCLNILKCRKQIFAACKSGKYVFCGKTIVATN-83’ (Table- 3). We also found the SVM based MHCII-IAb peptide regions, 171- PLAYYQNVK, 95- RTEAFIRTK, 90- DWTFKRTEA, 37- KAFYDANKT, (optimal score is 1.059); MHCII-IAd peptide regions, 159- LSSMTGLAP, 68- KSGKYVFCG, 11- KIKELLAGG, 49- TFTNCLNIL, (optimal score is 0.702); MHCII-IAg7 peptide regions , 128- PLVAAYGLN, 127- LPLVAAYGL, 18- GGSADVEIE, 39- FYDANKTLE, (optimal score is 1.551); and MHCIIRT1. B peptide regions, 49- TFTNCLNIL, 247- DKAFSASLS, 110- KSKNDAAKQ, 92- TFKRTEAFI , (optimal score is 1.092) which represented predicted binders from nucleocapsid protein (Table-2). The average propensity for the nucleocapsid protein is found to be above 1.027 (Figure- 5). All residues having above 1.0 propensity are always potentially antigenic (table-3). The predicted segments in nucleocapsid protein are ‘126- ELPLVAAYGLNVPASFDTCALRMMLCIGGPLPLLSSM- 162 and 50 FTNCLNILKCRKQIFAACKSGKYVFCGKTIVATN- 83’. Fragment identified through this approach tend to be high-efficiency binders, which is a larger percentage of their atoms are directly involved in binding as compared to larger molecules.
Future Perspectives
This method will be useful in cellular immunology, Vaccine design, immunodiagnostics, immunotherapeutics and molecular understanding of autoimmune susceptibility. Nucleocapsid protein sequence of Groundnut bud necrosis virus (GBNV) involved multiple antigenic components to direct and empower the immune system to protect the host from the nucleocapsid. MHC molecules are cell surface proteins, which take active part in host immune reactions and involvement of MHC class in response to almost all antigens and it give effects on specific sites. Predicted MHC binding regions acts like red flags for antigen specific and generate immune response against the parent antigen. So a small fragment of antigen can induce immune response against whole antigen. The method integrates prediction of peptide MHC class binding; proteosomal C terminal cleavage and TAP transport efficiency. This theme is implemented in designing subunit and synthetic peptide vaccines.