Entomology, Ornithology & Herpetology: Current Research
Open Access
ISSN: 2161-0983
ISSN: 2161-0983
Research Article - (2022)Volume 11, Issue 1
Identification of Chrysomya megacephala larva is simple and cheap using traditional microscopy techniques. Nevertheless, it may be hampered if the larvae come with incomplete morphological characteristics or are too immature for microscopic identification. Alternatively, the application of a molecular approach for the identification of these samples can be achieved without the need for taxonomy experts. Previously, mitochondrial DNA, Cytochrome Oxidase I (COI) gene is the most extensively used in the molecular study for the detection of Ch. megacephala blowfly. Toss date, this gene is reported to be least useful for identification, since several other fly species are found to be closely related and may cause erroneous interpretation during the identification process. Thus, this study aimed for a new and novel target gene called Ch. megacephala Gustatory receptor 1 gene (CmegGr1) which was never been used for identification before. The current study uses a molecular and bioinformatic technique to detect the presence of CmegGr1 in Ch. megacephala and its evolutionary relationship with other forensically fly species. The tissue and species specificity of this gene were also determined. The third instar larvae of Ch. megacephala and eight other forensically important fly species were obtained from two sources; the rabbit carcasses and the Forensic Entomology Unit collection. The DNAs were extracted from these larvae, followed by amplification of the CmegGr1 using Polymerase Chain Reaction (PCR). The resulting sequences were subjected to phylogenetic analysis. A 209 bp of the CmegGr1 gene was successfully amplified from 30 samples of Ch. megacephala with a sensitivity and specificity of 80% and 100%, respectively. CmegGr1 gene demonstrated a high specificity as none of the non-Ch. megacephala species were amplified.
Forensic larvae; Gustatory receptor 1 gene; Chrysomya megacephala; DNA identification; Phylogenetic study
Blowflies play an important role in estimating minimum postmortem interval; with several species found in human corpses. In Malaysia, the most prevalent calliphorid species re-covered from the corpses were Chrysomya megacephala [1] (Diptera: Calliphoridae), followed by Chrysomya rufifacies [2] (Diptera: Calliphoridae) [3]. As a significant number of Ch. megacephala was observed at the early stages of decomposition, it is one of the most useful species in assisting Malaysian entomologists. On top of that, the correct identification of species is a vital step in forensic investigation. The traditional identification of blowfly species in the field was based on morphological techniques that compared different morphologies of the adult or matured larvae across a wide range of morphological keys of forensic flies’ species. However, the identification and detection of this species could be difficult as the morphology of immature larvae has similarities with other species. This problem can be overcome by rearing the immature larvae until they are fully grown. However, in some cases, the larvae may end up dying, hindering the identification process.
A significant number of studies looking towards a molecular approach in identifying these flies have been done; mainly through comparison of known fly DNA sequences [4]. Studies have suggested the potential of a “barcoding region” within the mitochondrial DNA, Cytochrome Oxidase I (COI) gene, as a universal marker for molecular identification of the forensically important Diptera [5]. The usage of the COI gene has been established as a useful molecular marker in identifying blowfly’s species in Africa, Australia, Belgium, England, Germany, Lebanon as well as Malaysia [5-10]. However, over the years, the COI region was found to be inefficient in identifying Ch. megacephala, with recent studies demonstrating a possible hybridization and an incomplete lineage sorting between this species and Chrysomya saffranea [1]. As such, finding a new marker gene for this genus is crucial to avoid misinterpretations in DNA identification
The blowfly has an instinct in which it becomes attracted by the odour of a decaying corpse, reaching within minutes to lay eggs [11- 15]. A gustatory gene in Ch. megacephala, which was classified under the chemoreceptor’s superfamily, is usually involved in the sense of taste that corresponds to feeding behaviour, initiation of innate sexual responses as well as reproductive responses [15]. According, the Ch. megacephala Gustatory receptor 1 gene (CmegGr1) is highly expressed in the antennae and proboscises with maxillary palps in the adult fly. As such, this could be a promising target that would assist forensic scientists in the identification process of this species. The present study aimed to determine the potential of the CmegGr1 gene as a suitable marker in the identification of Ch. megacephala larvae. This study was done by comparing the availability of this gene in a group of Ch. megacephala larvae and several forensic important larvae species.
Sample collections
The third instar of Ch. megacephala larvae were collected from a simulation of rabbit carcasses at a forensic simulation site, Universiti Kebangsaan Malaysia (UKM), Selangor, Malaysia. The use of rabbit carcasses was approved by the UKM Ethics Committee (Approval No: PARAST/PP/2019/SYAMSA/30-JAN.2019-DEC.-2019-AR- CAT2.). The specimens were collected and directly preserved in 70% ethanol. The collected samples were stored at 4°C in the Entomology Laboratory, Faculty of Medicine UKM. All posterior and anterior spiracles were stored as vouchers.
DNA extraction
The genomic DNA was extracted from 30 pool samples of Ch. megacephala. Approximately, 25 mg of body tissue samples were used for each extraction and all the extracted DNA were stored at -20°C. The genomic DNA was extracted using QIAamp DNA Mini Tissue Kit (Qiagen, Germany) according to the manufacturer’s instructions with some modifications. After overnight incubation in ATL buffer, all the samples were grounded using sterile 1.5 ml tube plastic pestles. Following this, the samples were incubated for 5 minutes at 56°C. The DNA was then eluted with 200 μL of AE buffer and again after standing for 5 minutes in the AE buffer. The purity of the extracted DNA was quantified using a Nanodrop 2000c (Thermo Scientific®, USA) at an absorbance wavelength of A260/A280.
Polymerase chain reaction
The purified DNA was subjected to PCR amplification of CmegGr1 gene following the protocol outlined. The target fragments were amplified with the primer pairs CmegGr1-F/CmegGr1-B for CmegGr1 DNA fragment size of approximately 209 bp. The PCR mixtures contained ~150 ng template DNA, 1 unit of Taq DNA polymerase (Promega, USA), 1 x PCR reaction buffer (Promega, USA), 1.5 mM MgCl2 (Promega, USA), 200 μM of each dNTPs (Promega, USA), 0.4 μM of each forward and reverse primer and ddH2O to total up the mixture to 50 μL per reaction. The PCR was performed using an Eppendorf Mastercycler Pro Thermal Cycler with annealing temperature at 50°C for 1 minute and 30 seconds. The amplified products were visualized by electrophoresis using a 2% agarose gel at 180V for 30 minutes (Table 1).
| Primer | Sequences (5’-3’) | Temperature |
|---|---|---|
| CmerGr1-F | CACCACTTAAGATACCTCCT | Pre-denaturation: 94°C [5 minutes] |
| CmegGr1-B | TACGAGCAAACTTTTGGTAG |
|
Table 1: The sequenced primer used in this study and the temperature set for termacycler machine.
Sequence analysis
Samples that showed positive results were further sent for sequencing analysis using the Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) after purification using Hiyield Plus Gel/PCR Mini Kit (Real Biotech Corp, Taiwan). Gene sequence analysis and database comparisons were performed by the BLAST program. Nucleic acid sequences of nine additional insect species were retrieved from GenBank (Table 2).
| GenBank ID | Species | Country |
|---|---|---|
| MH750487.1 | Drosophila mojavensis | United States of America |
| MH750458.1 | Drosophila arizonnae | United States of America |
| MH750483.1 | Drosophila navojoa | United States of America |
| AB042625.2 | Drosophila melanogaster | Japan |
| KR674136.1 | Athetis dissimilis | China |
| XM013241847.1 | Stomoxys calcitrans | United States of America |
| JQ36177.1 | Musca domestica | United States of America |
| XM005189934.2 | Musca domestica | United States of America |
| KJ702098.1 | Calliphora stygia | Australia |
| XM023443869 | Lucilia cuprina | United States of America |
| JQ365174.1 | Chrysoma megacephala | China |
Table 2: GenBank ID used for sequence analysis.
All sequenced were aligned and trimmed using the BioEdit Sequence Alignment Editor Version 7.2.6 (Hall, 1999). Samples and database sequences were used to construct a phylogenetic tree to illustrate the relationships of each gene between species trees. The evolutionary tree was inferred using the Maximum-likelihood method using 1000 bootstrap.
Specificity test
For the specificity test, the PCR was performed on the other species of larvae using a similar protocol used for CmegGr1. DNA isolated from Ch. megacephala was used as a positive control. The non-Ch. megacephala selected was based on the availability of sources from our laboratory collections. The species available are Chrysomya rufifacies; Chrysomya nigripes Aubertin, Chrysomya villeneuvi Patton, Sarcophaga spp., Synthesiomyia nudiseta Wulp, Hypopygiopsis violacea; Hemipyrellia ligurriens (Wiedemann); and Musca domestica Linnaeus.
Detection of CmegGr1 from larvae
From the 30 pools of Ch. megacephala larvae, 80% (24 out of 30) of samples detected the CmegGr1 gene (Figure 1). The DNA sequence analysis revealed that the extracted sample from the larvae has DNA similarities of 96-98% with the adult CmegGr1 gene (Figure 2A). Further investigations showed that the phylogenetic tree of CmegGr1 has close characteristics with Calliphora stygia’s and Lucilia cuprina’s 21a gustatory receptors (Gr21a) as shown in Figure 2B.
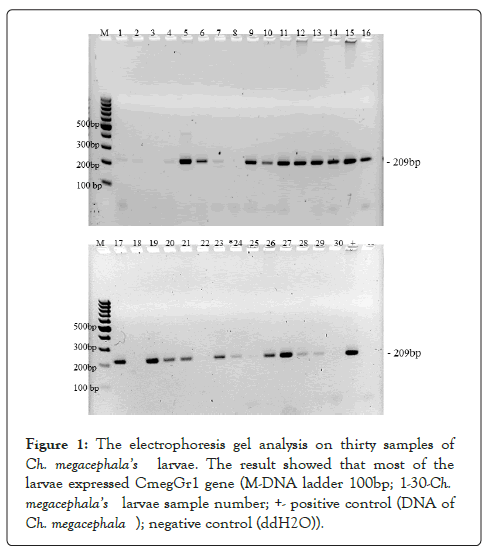
Figure 1: The electrophoresis gel analysis on thirty samples of Table 2: GenBank ID used for sequence analysis. Ch. megacephala’s larvae. The result showed that most of the larvae expressed CmegGr1 gene (M-DNA ladder 100bp; 1-30-Ch. megacephala’s larvae sample number; +- positive control (DNA of Ch. megacephala ); negative control (ddH2O)).
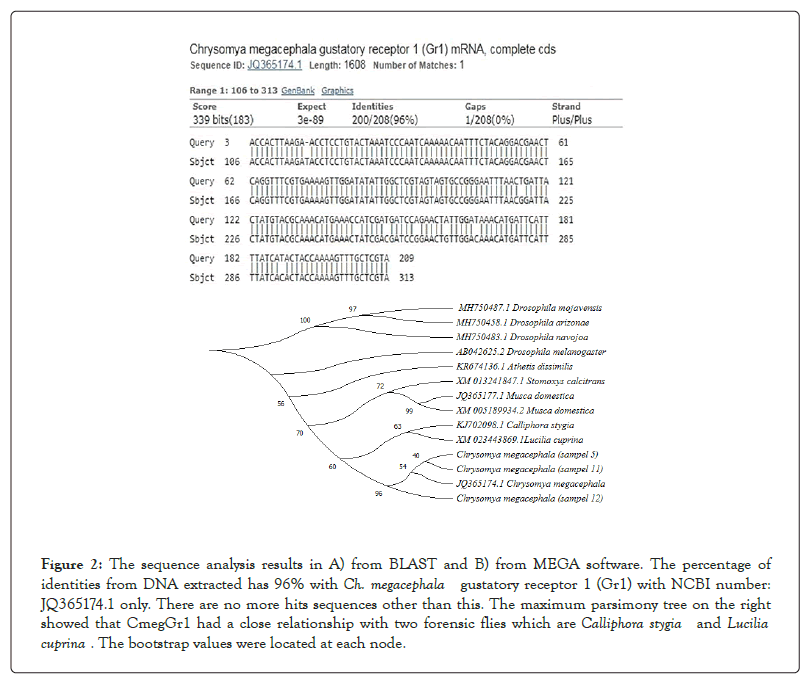
Figure 2: The sequence analysis results in A) from BLAST and B) from MEGA software. The percentage of identities from DNA extracted has 96% with Ch. megacephala gustatory receptor 1 (Gr1) with NCBI number: JQ365174.1 only. There are no more hits sequences other than this. The maximum parsimony tree on the right showed that CmegGr1 had a close relationship with two forensic flies which are Calliphora stygia and Lucilia cuprina . The bootstrap values were located at each node.
Illustrates the specificity results of CmegGr1 towards Ch. megacephala species only. CmegGr1 is exclusively present in Ch. megacephala samples but not in other necrophagous forensic larvae. All samples from Ch. rufifacies, Ch. nigripes, Ch. villeneuvi, Sarcophaga sp., Synthesiomyia nudiseta, Hypopygiopsis violacea, Hemipyrellia ligurriens and Musca domestica were found to be absent with CmegGr1 (Figure 3).

Figure 3: The electrophoresis gel analysis of CmegGr1. Specificity test using different species of forensic larvae. (M- DNA ladder 100bp; CM- Ch. megacephala ; CR-Ch. rufifacies; CN-Ch. nigripes; CV- Ch. villeneuvi; S-Sarcophaga sp.; SN-Synthesiomyia nudiseta ; HIPO-Hypopygiopsis violacea; HL-Hemipyrellia ligurriens; MD-Musca domestica; +-positive control (DNA of Ch. megacephala ); -- -negative control (ddH2O)).
In the context of using molecular techniques for identifying necrophagous larvae, it is crucial to choose the appropriate DNA target that could facilitate and complement the morphology-based species identification. It is undeniable that a reduced ability to recognize markers could lead to undesirable outcomes, especially in estimating PMI. This study focused on identifying new gene markers as one of the alternatives to current barcoding markers. In general, the COI gene, which consists of ≈ 1588 bp nucleotides, is the most common target amplicon used for molecular identification of the Ch. megacephala [16,17]. However, previous studies found that the COI gene was not specific towards the detection of Ch. megacephala [1,10]. Although this region has been used extensively, the lack of amplification specificity against other carrion-breeding blowflies such as Chrysomya pinguis, Chrysomya bezziana, Chrysomya pacific and Chrysomya chani highlights the shortcoming of this sequence in exclusively identifying Ch. megacephala [10,18,19]. As such, the above reasoning shows the instances where DNA barcoding utilizing the COI gene failed due to a lack of variability among different blowfly species, despite the inclusion of additional gene markers. Despite using a DNA barcoding technique, the identification method used in the present study was developed by amplifying a single DNA target that focuses on a single species only. The set of primers was designed using available sequence data. As such, this primer would only be able to attach to species that had the gene target. In line with the above, the detection of CmegGr1 as a specific genetic marker for Ch. megacephala could be used to accurately identify this fly from a sample containing a large species of forensic flies. The PCR result for the CmegGr1 gene showed a high specificity towards Ch. megacephala, suggesting its potential to become one of the genetic markers for this species. The detecting of the gene is noticeably less sensitive when using the conventional PCR method as it originates from nuclear markers which only have a few copies within the blowfly’s genome [13]. Fortunately, this shortcoming can be circumvented by utilizing methods such as real-time PCR or microarrays, which are known to possess a higher sensitivity.
From the result of the phylogenetic relationship of CmegGr1 with other Dipteran species, the 209bp of CmegGr1 gene could differentiate Ch. megacephala from other species as the sequence did not hybridize with gustatory receptors from other insects. Subsequently, the result from phylogenetic analysis found that the closest molecular characteristics of CmegGr1 were connected to Calliphora stygia’s and Lucilia cuprina’s 21a gustatory receptors namely CstyGr21a and LcupGr21a. Both flies can also be found at forensic sites. The molecular analysis by [20] also found that CmegGr1 has connections in molecular characteristics with CstyGr21a and gustatory gene from Drosophila melanogaster, which called DmelGr21a. These receptors were found to be important carbon dioxide receptors. This study proposed that CmegGr1 might function as one of the important receptors to detect volatile organic compounds for larvae Ch. megacephala. Another study by [20] also determined CmegGr1 gene as one of the carbon dioxide receptors, extracted from the adult Ch. megacephala. According to [21], carbon dioxide is one type of odorant and it is cuticular hydrocarbon gas. It is very volatile and important as long-distance cues for carrion detection [22]. Interestingly, carbon dioxide is the main gas product produced by the carcasses followed by hydrogen with 80% and 10% respectively at a constant temperature of 25°C [23].
Nonetheless, this study also deduced that CmegGr1 has a very specific molecular characteristic to Ch. megacephala and was not found from other forensic larvae. The amplification of CmegGr1 gave no result to the other eight forensic larvae available from our laboratory. The reason behind this is not because they do not have carbon dioxide receptors but the partial CmegGr1 that was chosen is very specific to Ch. megacephala only. The location of the amplicon was at the N-terminal of the protein and it is located at intracellular [20]. As the location of the sequences is not at transmembrane, the amino acids can easily adapt to the species selective preference. According to [24], some insects were born without carbon dioxide receptors such as honeybee, parasitoid wasp, human louse, pea aphid, water flea and blacklegged tick and some insects have carbon dioxide receptors such as Anopheles gambiae, Aedes aegypti and Culex pipiens. The behaviour of these insects that attracted either humans or corpses was paralleled to the presence of carbon dioxide receptors. The same study also showed that the N-terminal of Gr21a of 12 different species of Drosophilla spp. had distinct characteristics as they were found to be different from one to another. This adaptation and evolution process of carbon dioxide receptors was mentioned by [15] found that the sweet taste and carbon dioxide receptors have evolved across the species but at a very low rate, whereas bitter taste members evolve rapidly. Other than that, they also supported that most GR families are unique and species- specific. Therefore, these phenomena most probally showing that CmegGr1 has its unique molecular characteristics. This might be the key to the high specificity of Ch. megacephala and suggesting that this gene is suitable for identification of this species. Generally, the olfactory organs for immature larvae were found primarily at the terminal organ and antennal lobe (also called the dorsal organ). As had mentioned by [25], the detection of food in larvae is mediated by chemosensors that are located at a bilaterally symmetric anterior structure called antenna-maxillary complex which comprises these two organs. According to [26], in Drosophila melanogaster larvae, the terminal organ was mainly to detect soluble and volatile cues and comprised gustatory neurons whereas the dorsal organ was the main organ to detect chemicals and it has a lot of olfactory neurons. A study by [27] found that there are basiconic sensilla was found at the anal segment for Ch. megacephala larvae, which is also found greatly at the antenna. According to [28], the odorant receptors in fly appear in basiconic, intermediate and in trichoid sensilla which differ from receptors for ions. [25] had also found one type of gustatory in Drosophila spp. larvae namely Gr2B1 that was found to be expressed in the ventral pit. This receptor is related to one neuron in the terminal organ, two neurons in the dorsal organ and uniquely to a single bilaterally systematic neuron in each thoracic hemi segment. The high specificity of CmegGr1 may be used as a new identification method for Ch. megacephala by embedding the sequence into a microarray, using field samples originating from the immature larvae [29-31].
The sequence alignment of gustatory receptors of Ch. megacephala showed that the CmegGr1 and CmegGr2 amino acid sequences had different lengths in their N- and C- terminal regions compared to other insect species, thus providing distinguishing points for Ch. megacephala against other insects. Overall, the gustatory gene showed high specificity in insect identification through molecular detection methods and has the potential to be a reliable marker for genetic analyzers in the fieldwork. The specificity and sensitivity of the DNA identification of Ch. megacephala using CmegGr1 are beneficial in forensic investigations. The uniqueness of this gene and its ability to be expressed at the very immature stages enables it to be further developed as a diagnostic marker to help in identifying Ch. megacephala at forensic sites. This study could provide validation for future studies to reinvestigate similar genes on other species.
We also want to express our gratitude to Dr. Wong Kon Ken from the Department of Microbiology, UKM for assisting in perming the phylogenetic analysis and guiding the author to interpret the results.
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Citation: Ghazali SNA, Osman E, Othman H, Abdullah SR (2022) Potential Application of Gustatory Receptor 1 (CmegGr1) Gene as a Molecular Marker for Identification of Chrysomya megacephala (Diptera: Calliphoridae). Entomol Ornithol Herpetol. 11: 268.
Received: 04-Jan-2022, Manuscript No. Eohcr-21-15390; Editor assigned: 10-Jan-2022, Pre QC No. Eohcr-21-15390 (PQ); Reviewed: 24-Jan-2022, QC No. Eohcr-21-15390; Revised: 28-Jan-2022, Manuscript No. Eohcr-21-15390; Published: 04-Feb-2022 , DOI: 10.35248/2161-0983.22.11.268
Copyright: © 2022 Ghazali SNA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.