Immunome Research
Open Access
ISSN: 1745-7580
ISSN: 1745-7580
Research Article - (2022)Volume 18, Issue 2
Background: The objective of this study was to study the benefit of the rapid antigenic detection of SARS-CoV-2 and to demonstrate the contribution of this technique compared to real-time RT-PCR.
Methods: The SARS-CoV-2 nucleocapsid N antigen rapid diagnostic test (Standard Covid-19 Ag Test, SD Biosensor) was performed on 49 patients. Real-time RT-PCR testing was performed only in 12 patients.
Results: Nasopharyngeal swabs were taken from subjects whose mean age was 35 years (range 23-68 years) and who presented one of the following symptoms: dry cough (30.61%), chest tightness (28 %), fever (28%), headache (24.48%), asthenia (22.44%) and diarrhea in only 14.28%. The time between the onset of symptoms and the completion of the test ranged from 0 to 2 days. Of all rapid tests performed, 35 (71.42%) were negative and 14 (28.57%) were positive. Of the samples tested, 44 came from different IMKO departments. RT-PCR was performed in 8 patients whose rapid tests were negative and gave a positive result in 2 cases.
Conclusion: The detection of SARS-CoV-2 should be evaluated and compared to the standard RT-PCR technique, which often offers significantly better sensitivity. It is necessary to carry out large studies to better understand the issue of potential SARS-CoV-2 recurrence in COVID-19 patients.
Rapid antigen test; Diagnosis; COVID-19; SARS-CoV-2
In the late December 2019, a novel virus called Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), also known as 2019 novel coronavirus (2019-nCoV), was reported with an unidentified source [1]. The genomic sequence of this newly emerged virus is highly similar to that of severe acute respiratory syndrome coronavirus (SARS-CoV) with a 79.6% sequence identity [2]. This causes symptoms such as cough and fever, severe pneumonia, and death. The WHO reported that more than 280 million cases of COVID-19, including approximately 5, 4 million deaths, have occurred as of December 2021 (https://covid19.who.int/).
It has been reasoned that lung epithelium is the primary target of the virus. The Receptor-Binding Domain (RBD) of SARS-CoV-2 S-protein determines the entry of the virus into human host cells through the Angiotensin Converting Enzyme 2 (ACE2) receptor, and this mechanism is similar to the original SARS-CoV which shares significant homology in the RBD of S-protein [3].
Typical presentation of symptomatic COVID-19 includes fever, cough and fatigue, though other less frequent and atypical symptoms has also been described. Symptoms of COVID-19 have been enumerated in Figure 1 [3]. Severe COVID-19 has been defined as presence of tachypnea ( ≥ 30 breaths/min), oxygen saturation ≤ 93% at rest, >50% lung involvement on imaging or PaO2/FiO2 ratio<300 mm Hg, while critical disease has been defined as respiratory failure requiring mechanical ventilation, septic shock, or other organ dysfunction or failure requiring intensive care support [4]. The risk of severe and critical disease is highest in the elderly (>60 years) and those with underlying conditions like diabetes, hypertension, obesity, malignancy, chronic respiratory disease, cardiovascular disease and liver disease [5]. One study has suggested that children of all ages can get infected, and even though the majority of infections in children are asymptomatic or mild, outcomes can be poor in young children, especially infants with a 7%–10% incidence of severe and critical disease [6].
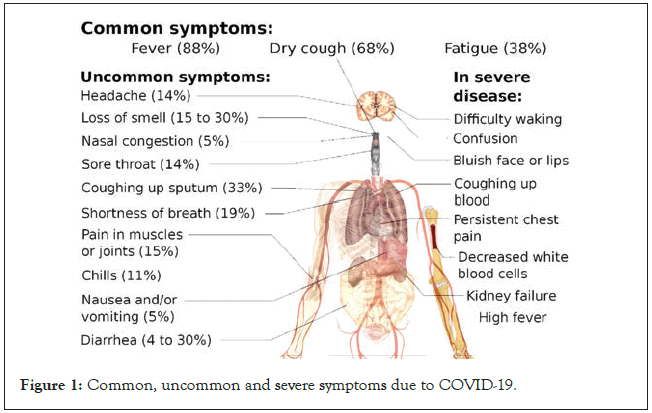
Figure 1: Common, uncommon and severe symptoms due to COVID-19.
The gold standard for COVID-19 diagnosis is reverse transcription- quantitative PCR (RT-qPCR) using nasopharyngeal (N) swabs, throat (T) swabs, or saliva [7]. RT-qPCR kits that do not require viral RNA extraction and high-throughput RT-qPCR systems have been developed. Although such tests are widely utilized in public health laboratories and large well-equipped hospitals, they are unavailable in local clinics where patients who suspect they have COVID-19 often goes first. Therefore, specimens need to be transported to and examined at sites that have RT-qPCR capability, which delays the test result and increases the anxiety of the suspected COVID-19 patients. To improve this situation, Rapid Antigen Tests (RATs) for COVID-19, which does not require specific and expensive machinery, have been approved for clinical use in Japan and other countries, and the sensitivity of these tests has been compared with that of several kinds of RT-qPCR [8–14].
Although these RATs might be useful for the identification of COVID-19 patients in local clinics, their sensitivity is important in determining usage strategies. Here, the objective of this work was to study the interest of the rapid antigenic detection of SARS- CoV-2 and examined the sensitivity of RATs available in the Manouba governorate in Tunisia between January and September 2021 for the detection of isolated viruses. We also evaluated their effectiveness and Highlight the contribution of this technique compared to real-time RT-PCR.
Rapid antigen tests
The SARS-CoV-2 N core antigen rapid diagnostic test (Standard Covid-19 Ag Test, SD Biosensor) [15] was performed in 49 patients. The SARS-CoV-2 Antigen Rapid Detection Test is a chromatographic immunoassay for the detection of SARS- CoV-2 core (N) antigen in the respiratory tract. The performance characteristics of the SARS-CoV-2 antigen rapid detection assay should be evaluated and compared to the standard real-time Reverse Transcription Polymerase Chain Reaction (RT-PCR) for the diagnosis of COVID-19 cases.
Reverse transcription polymerase chain reaction
The real-time Reverse Transcription Polymerase Chain Reaction (RT-PCR) assay was performed only in 12 patients using WANTAI transcriptase reverse real-time assays RT-PCR that target the Sarbecovirus envelope (E) gene, and RNA dependent RNA polymerase (RdRp) and the core gene (N) of SARS-CoV-2. SARS-COV-2 real-time RT-PCR (Reverse Transcription - Polymerase Chain Reaction) is a test based on the principle of multiplexed RT-PCR using specific primers and fluorescent probes.
This test is used to detect SARS-COV-2 nucleic acid in nasopharyngeal, or pharyngeal and sputum samples. Alpha Prep TM VIRAL DNA/RNA Extraction kit (Model: VDR-B096V) was used for extraction of nucleic acids from various cell types. The extracted nucleic acids are applicable PCR, real-time PCR, and enzymatic reaction etc.
Preparation of plate
Remove the sealing cover and add 200 μl of sample and 20 μl of proteinase K to the 1st or 7th well in the plate. Insert the plate correctly in the instrument (GenMagBio RNA extractor reagent kit with reliable quality).
Extraction of nucleic acids
After inserting the plate, close the door of the instrument and enter the correct program. When operation is finished, remove the plate from the instrument and then transfer 80 μl of extracted nucleic acids 6th or 12th to the 1.5 ml tube. Store the extracted nucleic acids at -20°C or -70°C. Discard the used 8- strip and the plate.
Amplification
The SARS-COV-2 RNA sequences are amplified by the presence of specific primers. These were designed to amplify in vivo a specific region of nucleic acid: the structural genes N, RDRP, E, and S. The time RT-PCR were evaluated according to the procedures described in the manufacturers’ instructions.
WANTAI Test principle
This kit is a qualitative, real-time fluorescent PCR in which specific primers and fluorescent probes are designed to detect the highly conservative regions of the ORF1ab and N genes of the virus. This kit has integrated quality control (IC, human housekeeping gene) intended for monitoring of the test run.
Reagents preparation
Step 1- Prepare the reagents: Open the kit and remove the components from the box. Thaw at room temperature; shake to mix for 1 minute then centrifuge immediately. Place the RT-PCR master mix, Mn (OAc)2, primers and probes at 2~8°C to refrigerate for later use.
Step 2- Prepare the PCR reaction mix: One test requires 30 L of PCR reaction mix. Depending on how many specimens will be tested, mix the required volumes of reagents as per the Table 1 below. Centrifuge intermediately after mixing thoroughly. It is advised to prepare one additional test reagent each time to prevent the loss of reaction mix due to splitting.
| Component | Volume per 1 réaction (µl) |
|---|---|
| RT-PCR Master mix | 25 |
| Primer probe | 2.5 |
| Mn (Mn (OAc)2 | 2.5 |
| Total | 30 |
Step 3 - Transfer to PCR reaction tube: Pipette 30 L of the PCR reaction mix into a PCR reaction tube (choose a PCR reaction tube compatible with the extractor instrument).
Step 4 - Add the RNA template: Add 10L of RNA template or controls to the PCR amplification tube. Close the tube and centrifuge instantly. Transfer to the amplification and analysis area for PCR amplification. (This kit does not contain RNA extraction reagents. Suggested extraction kits and equipment: Beijing Wantai, GenMagBio commercialized RNA extractor reagent kit with reliable quality (Table 2).
| S.no | Steps | Température | Time | Cycles |
|---|---|---|---|---|
| 1 | UDG enzyme action | 37°C | 2 min | 1 |
| 2 | RNA dénaturation | 90°C | 30 secs | 1 |
| 3 | RNA reverse transcription | 61°C | 15 min | 1 |
| 4 | Dénaturation | 95°C | 3 secs | |
| Annealing, fluorescence signal gathering | 60°C | 10 secs | 45 |
Amplification and analysis area
Place the PCR amplification tube into the RT-PCR instrument. Label to indicate the controls and sample positions. Select FAM for ORF1a gene, VIC /HEX for the N gene, and ROX for the IC. Set the PCR reactionmix volume to 40 Μl set the cycles according to the table below.
Nasopharyngeal swabs were taken from subjects with a mean age of 35 years (range 23-68 years). The time from start to lab test in suspected COVID-19 cases ranged from 0 to 2 days. The patients presented mainly one of the following symptoms: dry cough (30.61%), chest tightness (28%), fever (28%), headache (24.48%), asthenia (22.44%) and diarrhea. in only 14.28% (Figure 2). Of all rapid tests performed, 35 (71.42%) were negative and 14 (28.57%) were positive (Figure 3). Of the samples tested, 44 came from different IMKO departments. RT-PCR was performed in 8 patients whose rapid tests were negative and gave a positive result in 2 cases. The RT-PCR result of two samples had relatively Ct values as follows; ORF 1ab (FAM)=37.76, N (VIC)=41.65 and CI (ROX)=32.56 (WANTAI) (Table 3) (Figure 4).
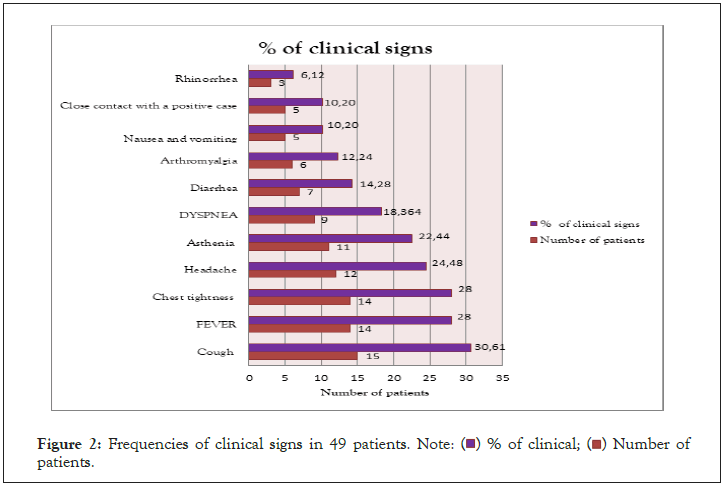
Figure 2: Frequencies of clinical signs in 49 patients. Note: ( ) % of clinical; (
) % of clinical; ( ) Number of
patients.
) Number of
patients.
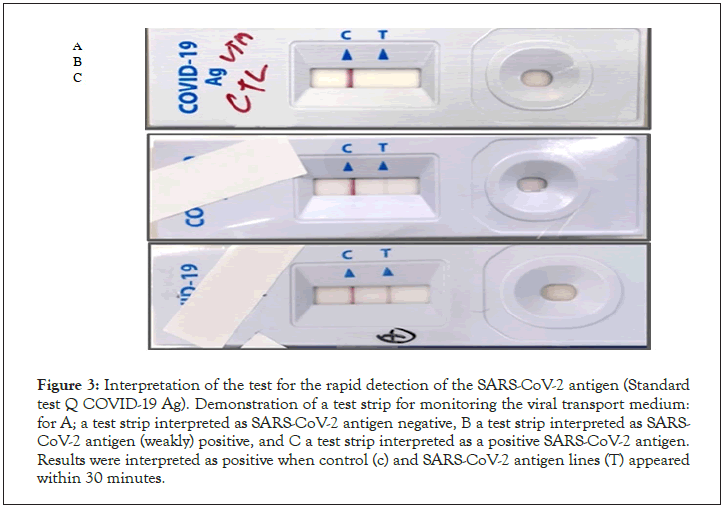
Figure 3: Interpretation of the test for the rapid detection of the SARS-CoV-2 antigen (Standard test Q COVID-19 Ag). Demonstration of a test strip for monitoring the viral transport medium: for A; a test strip interpreted as SARS-CoV-2 antigen negative, B a test strip interpreted as SARS- CoV-2 antigen (weakly) positive, and C a test strip interpreted as a positive SARS-CoV-2 antigen. Results were interpreted as positive when control (c) and SARS-CoV-2 antigen lines (T) appeared within 30 minutes.
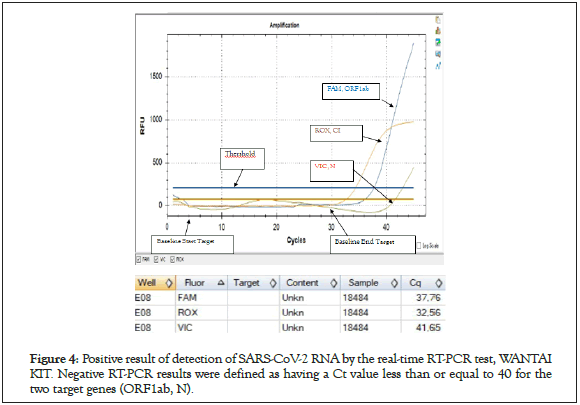
Figure 4: Positive result of detection of SARS-CoV-2 RNA by the real-time RT-PCR test, WANTAI KIT. Negative RT-PCR results were defined as having a Ct value less than or equal to 40 for the two target genes (ORF1ab, N).
| NO | FAM(ORF1ab) | VIC(N) | ROX | Result détermination and action |
|---|---|---|---|---|
| (IC) | ||||
1 |
Ct ≤ 40 | Ct ≤ 40 | / | SARS-COV-2 positive |
2 |
No Ct or Ct=45 | No Ct or Ct=45 | ≤ 35 | SARS-COV-2 negative |
3 |
Ct > 40 or No Ct | Ct < 45 | / | Re-extraction and retest are needed. During retest, if any target has a Ct<45, then it is judged as positive. If the two targets have no Ct (or Ct<45) and internal control has a Ct of ≤ 35, then it is judged as negative. If internal control is >35, then judge the results according to No. 5. |
4 |
Ct < 45 | Ct > 40 or No Ct | / | |
5 |
No Ct or Ct=45 | No Ct or Ct=45 | >35 or No Ct | Test is invalid. Re-extraction and retest are needed. If retest results are still >35 (or No Ct), then judge as Specimen inhibition. |
The recommended samples for these RATS are nasopharyngeal swabs not saliva. Other study found that some of the saliva samples from which SARS-CoV-2 was isolated gave negative results with all of the RATs [15]. Nonetheless, the RATs need to be improved with respect to their sensitivity.
Our results of clinical signs are similar with what has been reported in China (1). Several results [16,17] have shown a higher sensitivity of this SARS-CoV-2 antigen test (98.33% according to the Q Test COVID-19 Ag standard) compared to other rapid antigenic tests; 93.9% for the Ag 2019-nCoV test (Bio easy Biotechnology Co., Shenzhen, China), 50.0% by COVID-19 Ag Respi-Strip CORIS®, and 11.1 to 45.7% by BIOCREDIT COVID-19 Ag (Bio Vendor Research and Diagnostics).studies [18] have shown that this SARS- CoV-2 antigen detection kit could be recommended for patients at the onset of symptoms after the onset of symptoms where higher viral loads are anticipated. Other factors such as the type of clinical manifestation the time between onset of illness and use of the laboratory test tended to give false negative results. Our study confirmed that the perfect detection of SARS-CoV-2 should be evaluated and compared to the standard RT-PCR technique, which often offers significantly better sensitivity. The RT-PCR was performed in 8 patients, whose rapid tests were negative and gave a positive result in 2 cases, currently, contradictories studies found that there is a certain possibility of RT-PCR rendering false negative results, including due to the sampling procedure, sources of samples and the sensitivity/specificity of the nucleic acid test kit [19]. Indeed, a high false-negative rate (48/384, 12.5%) of RT-PCR results for SARS-CoV-2 detection was observed [20] 36 days vs. 21 days, p<0.001, compared to control group [21]. Nasal swab sampling rather than throat swabs and anal swabs for SARS-Cov-2 testing could reduce the false negative rate of nucleic acid tests [21]. Currently, due to underestimated proportion of patients with prolonged SARS-CoV-2 RNA conversion and high false negative rate of viral test results, recurrence of positive SARS-CoV-2 may experience from false negative RT-PCR results.
Alternatively, it cannot be excluded that truly negative discharged patients suffered reactivation or were re-infected with another SARS-CoV-2 strain. SARS-CoV-2 reactivation or re-infection will be a persistent and vexing problem. It is a major public health concern in terms of global morbidity and possibly mortality. We suggest conducting a genetic characterization of viruses to distinguish between reactivation and re-infection with SARS-CoV-2 in the context of confirmed recurrence during patient convalescence. In addition, SARS-CoV-2 RNA detection was found positive in anal swabs but negative in nasopharyngeal swabs for 42 days in an asymptomatic carrier, raising the question of a possible transmission via faecal-oral route [22,23] Serological tests could be used to facilitate identification of asymptomatic COVID-19 patients in the community [24]. Considering the significance of this ongoing global public health emergency, it is necessary to carry out large studies to better understand the issue of potential SARS- CoV-2 recurrence in COVID-19 patients.
Due to limited proportion of patients with prolonged SARS-CoV-2 RNA conversion and high false negative rate of viral test results, recurrence of positive SARS-CoV-2 may experience from false negative RT-PCR results. SARS-CoV-2 reactivation or re-infection will be a persistent and vexing problem. The detection of SARS-CoV-2 should be evaluated and compared to the standard RT- PCR technique, which often offers significantly better sensitivity. It is necessary to carry out large studies to better understand the issue of potential SARS-CoV-2 recurrence in COVID-19 patients. Conducting a genetic characterization of viruses to distinguish between reactivation and re-infection with SARS-CoV-2 in the context of confirmed recurrence during patient convalescence is required.
None of the authors of this paper has a financial or personal relationship with other people or organizations that could in appropriately influence or bias the content of the paper.
This work was supported by the Tunisian ministry of Health.
The laboratory is financed by the Ministry of Health according to a national strategy of diagnostic COVID 19.
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
Citation: Kilani H, Lamine YB, Kaoual S, Chatti L, Jemaa RB, Boubaker IBB et al. (2022) Place of Rapid Diagnostic Test in the COVID-19 Management Strategy in Tunisia. Immunome Res 18: 159.
Received: 06-Jul-2022, Manuscript No. IMR-22-18397; Editor assigned: 11-Jul-2022, Pre QC No. IMR-22-18397 (PQ); Reviewed: 25-Jul-2022, QC No. IMR-22-18397; Revised: 01-Aug-2022, Manuscript No. IMR-22-18397 (R); Published: 08-Aug-2022 , DOI: 10.35248/1745-7580.22.18.159
Copyright: © 2022 Kilani H, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.