Research Article - (2025)Volume 11, Issue 1
Phenotypic and Molecular Characterization of Multidrug-Resistant Uropathogenic Escherichia coli
Tonushyam Sonowal* and Shariff MAbstract
Background: Urinary Tract Infection (UTI) is the second most common infection after respiratory tract infections in the community. In hospitals, UTI is the most prevalent infection accounting for approximately 35% of all hospitalacquired illnesses. Uropathogenic E. coli (UPEC) accounts for 70% of all cases. Many MDR isolates of E. coli have been reported and a significant increase in rates makes it a difficult-to-treat pathogen.
Aim: Detection of production of beta-lactamases in Multidrug-Resistant (MDR) urinary isolates of Escherichia coli by phenotypic and molecular methods.
Materials and methods: Uropathogenic Escherichia coli (UPEC) isolates confirmed by MALDI-TOF were included in the study. The isolates were tested for antibiotic sensitivity to different antimicrobials and the production of beta-lactamases viz. In multi-drug resistant isolates, ESBL, MBL and AmpC were tested by both phenotypic and genotypic assays. Their clonality was determined by the RAPD.
Results: Out of a total of 1150 urinary samples, 122 yielded pathogens of which 55 were E. coli. Twenty-nine (52.7%) isolates that were multi-drug resistant were tested for the presence of beta-lactamases. All (29) isolates were positive for ESBL and 6 isolates also harbored MBL genes by PCR. None of the isolates produced AmpC. OXA (n=21) was the commonest followed by TEM and SHV. Among the MBLs, SPM, GIM, IMP and VIM were detected. Thirteen isolates had multiple types of beta-lactamase genes.
Conclusion: 53 percent of our E. coli isolates were MDR. All of them produced beta-lactamases. Hence, routine ESBL testing for uropathogens would be useful for all cases of UTI.
Keywords
Beta-lactamase; ESBL; Multidrug resistance; RAPD; Uropathogenic Escherichia coli
Introduction
Urinary Tract Infections (UTIs) are the most common infections mostly caused by gram-negative bacteria. Almost 150 million cases of UTIs per year are reported worldwide. According to the studies, Uropathogenic Escherichia coli (UPEC) is the most common cause of UTIs. UPEC isolates possess multiple virulence factors that promote colonization of the bacteria and infection in the urinary tract such as fimbrial, adhesins, afimbrial adhesin, toxins, siderophores and capsular polysaccharides. The clinical experiences have shown a high rate of antibiotic resistance among uropathogens [1]. Excessive use of antibiotics is the most important factor in the development of Multidrug Resistance (MDR) in UPEC isolates. Antibiotic resistance is a health concern causing more mortality and morbidity. The knowledge of the bacteria causing UTIs and their antimicrobial susceptibility is necessary for appropriate empirical therapy and prevention of the emergence of antibiotic resistance. The present study was conducted to examine the sensitivity of UPEC and the production of beta-lactamases in urinary isolates of Multidrug-Resistant (MDR) E. coli by phenotypic and molecular methods.
Materials and Methods
Study design
It is a retrospective study. Isolates present in our repository were included. Clinical and demographic details were obtained from the records. Urine specimens sent to the microbiology department were processed as per standard protocols [2]. The isolates, that were of significant counts, ≥ 104 colony-forming units/ml for catheter specimens or ≥ 105 colony-forming units/ ml for clean-catch specimens, were identified using the vitek cards and/or MALDI-TOFF (Bruker Daltonics) and were further tested for antimicrobial susceptibility and maintained in glycerol broth at -70°C until used for further testing.
Antibiotic susceptibility testing
The susceptibility of isolates to various antibiotics was tested by Kirby Bauer’s disk diffusion method. Ceftazidime (30 μg), ceftazidime/clavulanate (30/10 μg), cefoxitin (30 μg), cefepime (30 μg), levofloxacin (5 μg), aztreonam (30 μg), gentamicin (10 μg), meropenem (10 μg), piperacillin (100 μg), piperacillin/ tazobactam (100/10 μg), tigecycline (15 μg), nitrofurantoin (300 μg), norfloxacin (10 μg) and co-trimoxazole (25 μg) were used. Briefly, the test organism was grown in mueller hinton broth and incubated at 37°C for 2-3 hours. The turbidity was adjusted to match 0.5 McFarland standards. A lawn culture of the organism was made in Mueller Hinton Agar (MHA) plates. Antibiotics-impregnated disks were placed on the surface of the plate and were incubated overnight at 37°C. The diameter of the zone of inhibition was recorded and interpreted as sensitive, intermediate or resistant as per the CLSI guidelines 2018. They were categorized as Multi-Drug Resistance (MDR), Extensive Drug-Resistance (XDR) and Pan-Drug Resistance (PDR). Out of 55 isolates, 25 isolates were MDR and 4 were XDR. They were further characterized for the presence of beta-lactamase genes [3].
Production of ESBL, MBL and AmpC
The E. coli isolates were screened to produce beta-lactamases and further confirmed as per CLSI guidelines 2018. Briefly, for ESBL production a 0.5 McFarland suspension of each isolate was spread on the MHA plate and disks of cephalosporin and cephalosporin/clavulanate were placed aseptically about 15 mm apart (edge-to-edge) and the cultures were incubated at 37°C overnight. A difference of ≥ 5 mm between the zone diameters of the two disks was confirmatory of ESBL production. The difference in the zone diameter is due to the inhibition of the β- lactamases by clavulanic acid.
Isolates were also tested for MBL production by combined disc test. Briefly, two meropenem disks (10 μg) were placed approximately 30 mm apart, on the surface of the MHA plate, inoculated with test organism and an appropriate amount of 0.5M EDTA solution was added to one of the disks to obtain a concentration of 750 μg. This was incubated overnight at 37°C and when there was an increase of ≥ 7 mm in the zone of inhibition around the meropenem+EDTA disk as compared to the meropenem disk alone it was considered as an MBL producer [4].
To confirm the isolates for AmpC production, an AmpC disc test was done. Briefly, a lawn culture of cefoxitin-sensitive E. coli (ATCC 25922) was prepared on an MHA plate and a cefoxitin disk (30 μg) was placed on the surface of the inoculated medium. A sterile plain disk of 6 mm diameter was placed next (almost touching) to the cefoxitin disk, moistened with 20 μl of sterile saline and inoculated with several colonies of the test organism. The plate was incubated overnight at 37°C. A flattening or indentation of the cefoxitin inhibition zone in the vicinity of the inoculated disc was considered a positive test.
Polymerase Chain Reaction (PCR)
DNA was extracted from the isolates using HiPurA™ bacterial genomic DNA purification kit (HiMedia, Catalog no. MB505-50PR) following the manufacturer's instructions. The extracted DNA was stored at -20°C till further use.
The E. coli isolates were tested for the presence of the genes that encode ESBLs, MBLs and AmpC by PCR. Different primers were used to amplify the genes and the amplification reaction was carried out by the methods described (Table 1).
| Genes tested | β-lactamases targeted | Sequence | Amplicon size (bp) | PCR cycle condition |
|---|---|---|---|---|
| Multiplex I TEM, SHV and OXA-1-like | TEM variants including TEM-1 and TEM-2 | CATTTCCGTGTCGCC CTTATTC CGTTCATCCATAGTT GCCTGAC |
800 | Initial denaturation at 94°C for 10 min; 30 cycles of 94°C for 40 s, 60°C for 40 s and 72°C for 1 min; final elongation step at 72°C for 7 min |
| Multiplex I TEM, SHV and OXA-1-like | SHV variants including SHV-1 | AGCCGCTTGAGCAA ATTAAAC ATCCCGCAGATAAA TCACCAC |
713 | |
| OXA-1, OXA-4 and OXA-30 | GGCACCAGATTCAA CTTTCAAG GACCCCAAGTTTCCT GTAAGTG |
564 | ||
| Multiplex VIM, IMP, GIM, SPM, SIM | VIM variants including VIM 1 and VIM-2 | GATGGTGTTTGG TC CATA CGAATGCGCAGC ACCAG |
390 | Initial denaturation at 94°C for 5 min; 36 cycles of 94°C for 30 s, 52°C for 40 s and 72°C for 50 sec. final elongation step. at 72°C for 5 min |
| Multiplex VIM, IMP, GIM, SPM, SIM | IMP variants except IMP-9, IMP-16, IMP 18, IMP-22 and IMP-25 | GGAATAGAGTGG CTTAAYTCTC CCAAACYACTAS GTTATCT |
188 | |
| GIM-1 | TCGACACACCTT GGTCTGAA AACTTCCAACTT TGCCATGC |
477 | ||
| SPM-1 | AAAATCTGGGTA CGCAAA CG ACATTATCCGCT GGAACAGG |
271 | ||
| SIM-1 | TACAAGGGATTC GGCATCG TAATGGCCTGTT CCCATGTG |
570 | ||
| MOXM | MOX-1, MOX-2, CMY-1, CMY-8 to CMY-11 |
GCTGCTCAAGGAG CACAGGAT CACATTGACATAG GTGTGGTGC |
520 | Initial denaturation at 94°C for 3 min; 35 cycles of 94°C for 30 s, 64°C for 30 s and 72°C for 1 min. final elongation step at 72°C for 7 min |
| CITM | LAT-1 to LAT-4, CMY-2 to CMY-7, BIL-1 |
TGGCCAGAACTGA CA GGCAAA TTTCTCCTGAACGT GGCTGGC |
462 | |
| DHAM | DHA-1, DHA-2 | AACTTTCACAGGT GTGCTGGGT CCGTACGCATACT GGCTT TGC |
405 | |
| ACCM | ACC | AACAGCCTCAGCA GCCGGTTA TTCGCCGCAATCAT CCCAGC |
346 | |
| EBCM | MIR-1T/ ACT-1 | TCGGTAAAGCCGA TGTTGCGG CTTCCACTGCGGCT GCCAGTT |
302 | |
| FOXM | FOX-1 to FOX-5b | AACATGGGGTATC AGGGAGATG CAAAGCGCGTAAC CGGATTGG |
190 |
Table 1: Various primers used for ESBL, MBL and AmpC PCR.
PCR amplification was carried out using PTC-100 Peltier Thermal Cycler (bio-rad).
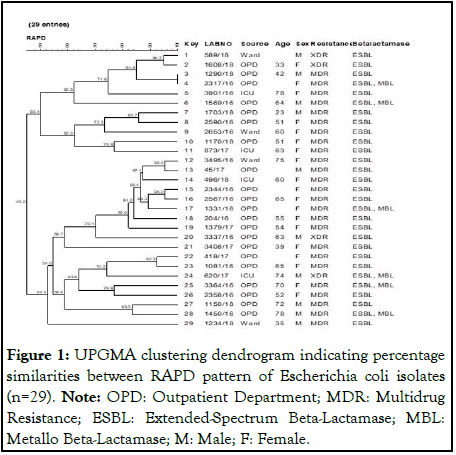
Amplified products were then separated by electrophoresis in 1.3% agarose gels containing Hi-SYBr safe gel stain (HiMedia, Catalog No. ML053). A 100 bp DNA ladder was used as a molecular weight marker (New England BioLabs Inc., Catalog No. N3231S) (Figure 1).
Figure 1: UPGMA clustering dendrogram indicating percentage similarities between RAPD pattern of Escherichia coli isolates (n=29). Note: OPD: Outpatient Department; MDR: Multidrug Resistance; ESBL: Extended-Spectrum Beta-Lactamase; MBL: Metallo Beta-Lactamase; M: Male; F: Female.
RAPD
All the 29 MDR E. coli isolates were typed by random amplification of polymorphic DNA using ERIC-2 primer (AAGTAAGTGACTGGGGTGAGCG). The reaction mixture was prepared in a final volume of 25 μl containing 2.5 μl of 10X PCR buffer, 0.5 μl of dNTPs, 0.5 μl of taq polymerase, 2 μl of DNA sample, 1 μl of 2.5 pmol ERIC-2 primer and nuclease-free water [5]. PCR amplification was carried out using BIO-RAD PTC-100 peltier thermal cycler under the following conditions: Denaturation at 94°C for 5 min, followed by 40 cycles each at 94°C for 1 min, annealing at 50°C for 1 min and extension at 72°C for 1 min with a final elongation step of 72°C for 10 min. PCR products were resolved in 1.3% agarose gel containing Hi- SYBr safe gel stain (HiMedia product code ML053-500 μl. A 100 bp DNA ladder was used as a molecular weight marker (New England BioLabs Inc., Catalog No. N3231S). The gels were analysed using GelCompar II software (Applied-Maths, Kortrijk, Belgium) and the dendrogram was generated using the unweighted pair group method with arithmetic mean (Figure 1).
Results
Study population and patient characteristics
A total of 1150 urinary samples, collected from the various patients attending the OPD and admitted in wards and ICUs were included. Patients of both sexes were included in the study. Most of the patients were female. The median age was 33 years. Most of the patients presented with fever and burning micturition at the time of sample collection [6].
Study outcome
Out of 1150 urinary samples, 122 samples yielded significant counts of pathogens. The majority were Escherichia coli (55) followed by Klebsiella species (24). These 55 Escherichia coli isolates belonged to 18 males and 37 females. Forty-two were from the outpatient department and 7 and 6 from wards and ICU respectively. A varying degree of resistance to antimicrobials was observed in the 55 isolates (Table 2).
| Antimicrobials | Resistant (n) | Sensitive (n) | Percentage resistance |
|---|---|---|---|
| G/Gentamicin | 24 | 31 | 43.6 |
| CA/Ceftazidime | 36 | 19 | 65.45 |
| CX/Cefoxitin | 24 | 31 | 43.6 |
| LE/Levofloxacin | 39 | 16 | 70.9 |
| PC/Piperacillin | 38 | 17 | 69 |
| PIT/Piperacillin- Tazobactam | 14 | 41 | 25.45 |
| Mr/Meropenem | 10 | 45 | 18.1 |
| AT/Aztreonam | 39 | 16 | 70.9 |
| CPM/Cefepime | 40 | 15 | 72.7 |
| NF/Nitrofurantoin | 9 | 46 | 16.3 |
| NT/Netilmicin | 8 | 47 | 14.54 |
| Na/Nalidixic Acid | 48 | 7 | 87.2 |
| NX/Norfloxacin | 36 | 19 | 65.4 |
| CO/Cotrimoxazole | 37 | 18 | 67.2 |
Table 2: Antibiogram of uropathogenic E. coli isolates (n=55).
Twenty-nine (52.7%) isolates were MDR, four of which showed Extensive Drug Resistance (XDR). MARI index of all 29 (72.4%) MDR isolates was ≥ 0.2. However, 19 (73.07%) of the 26 non-MDR isolates had a MARI of ≥ 0.2. All 29 MDR E. coli isolates were further tested for the presence of beta-lactamases. PCR was found to be more sensitive in detecting various betalactamase production when compared to phenotypic tests.
All 29 were positive for beta-lactamases on screening and 11 were confirmed to harbour Extended-Spectrum Beta-Lactamases (ESBL). However, all of them were positive for ESBL genes by PCR. OXA 1-like (21 (72.4%)) was the commonest gene followed by TEM (13 (44.8%)) and SHV (2 (6.8%)). Only two isolates were confirmed for MBL production by phenotypic methods, but six isolates showed the presence of MBL genes. Among the MBL genes, SPM (6 (100%)), GIM (4 (66.6%)), IMP (3 (50%)) and VIM (2 (33%)) were detected. Thirteen isolates showed multiple types of beta-lactamase genes [7]. None was positive for AmpC genes. Six isolates had both ESBL and MBL genes (Table 3).
| ESBL (n=29) with different genes | No of isolates (n) |
|---|---|
| OXA | 21 |
| TEM | 13 |
| SHV | 2 |
| MBL(n=7) showing different genes | |
| SPM | 6 |
| GIM | 4 |
| IMP | 3 |
| VIM | 2 |
| SIM | 0 |
Table 3: Distribution of ESBL, MBL and AmpC genes among the MDR E. coli isolates.
RAPD
The clonality of the 29 resistant isolates was determined by Randomly Amplified Polymorphic DNA (RAPD). The discriminatory index is 0.99 showing that RAPD was a good test for discrimination of clones. Among these 29 isolates, 24 clones were seen. Two isolates from a single clone showed 100% similarity. Both the isolates were from OPD and produced ESBLs. One of them produced MBL in addition. Four different clones with 2 isolates each were around 95% similar. The isolates from one of these clones were XDRs. Overall, there was not much difference observed between the isolates.
Discussion
Multi-drug resistance in uropathogenic Escherichia coli is an emerging serious public health problem that can lead to treatment failure. Twenty-nine (52.7%) isolates of E. coli were MDR. Low resistant rates, 15%, 16.6%, 18.8% and 25% were observed for netilmicin, nitrofurantoin, meropenem and piperacillin-tazobactam respectively. The carbapenem resistance is similar to ours in a study from India. However, a few studies from India and Nepal showed a very high resistance of about 60%-75%. In the present study, for most other antibiotics, the resistant rates were above fifty per cent and as high as 87% for nalidixic acid. This is like that observed in Bangladesh. In a study by Hasan et al. in a tertiary care Indian hospital the prevalence of MDR E. coli was about 52.9% similar to the present study. A study by Mathai et al. in southern India has also shown that 8.4% of commensal E. coli were MDR. Biofilm formation inside the bladder is an important factor that increases the selection of MDR strain leading to recurrent UTI. Studies from India have reported E. coli as one of the most common organisms causing UTIs. Various studies show that the prevalence of MDR UPEC isolates in developing countries is higher varying from 42% (China), 49.8% (Iran) and 68% in Pakistan or even 98% in Mexico. MDR bacteria, UPEC included, are responsible for almost 25 thousand deaths every year in Europe alone and for 700,000 deaths worldwide [8].
MARI is the index which identifies high-risk sources, like contamination of food, water and in recent times antibiotic consumption, of drug-resistant bacteria causing infections. In the present study, the percentage of isolates having a MARI index of >2 was 72.7 and most of these patients 42 (76%) were outpatients. The higher risk could be due to the widespread availability of antimicrobials without prescription, selfmedication, non-compliance of dosage and irrational use across, agriculture, poultry and cattle leading to greater exposure to multidrug-resistant bacteria in the community.
In our study, all the MDR E. coli isolates (29) were positive for ESBL and 6 of them were also positive for MBLs by PCR. OXA was the commonest followed by TEM and SHV. The results are discordant with studies where 87.1% TEM, followed by 70.6% SHV, SHV (43.1%) exceeded TEM (35.2%), CTX M (28.8%) exceeded SHV (13.7%) and CTX M (71.4% in E. coli exceeded TEM (55.1%). This could be because these studies are comparatively old and the trend has changed since then. Several other studies that have been performed throughout the world also showed variable results. In the years before 2000, TEM was the commonest of ESBLs which was then replaced by CTXM gene in the Indian bacterial population. In urinary isolates, OXA was predominant. In the present study, CTXM was not tested. However, in some of our recent uropathogenic E. coli isolates, we have found CTXM 15 (unpublished data). These isolates are known to be associated with a clone lineage belonging to ST 131. All our isolates belonging to ST 131 harboured CTXM 15. However, some of the other CTXM 15 isolates belonged to different lineages. These spread via Mobile Genetic Elements (MGEs) leading to a rapid increase in MDRs and E. coli causing UTIs and BSIs. Among the MBLs, SPM was the most common followed by GIM IMP and VIM. However, in our recent isolates, NDM5 was predominant with very few TEMs. So, there is a constant evolution of the bacteria leading to different beta-lactamases becoming predominant due to the antimicrobials in circulation. The occurrence of more than one beta-lactamase within the same isolate has been detected in our study. Thirteen isolates showed multiple types of beta-lactamase genes. Multiple beta-lactamases can be produced in gramnegative species which occurs with high frequency in carbapenem-producing MDRs. This could be because of their wider dissemination due to mobile genetic elements. None of our isolates were positive for AmpC genes. However, in a recent study, 68% of isolates showed AmpC beta-lactamase. In the same study, the most prevalent gene was bla NDM (80%). bla NDM was not tested in the present study. However, recent isolates of uropathogenic E. coli from our laboratory showed a preponderance of bla NDM 5 (unpublished data). Apart from NDMs, bla OXA 47 and bla OXA-1 are associated with carbapenem resistance. We have tested for bla OXA-1 and found it to be 72.4% of the carbapenem-resistant isolates [9].
All 29 isolates were further typed by Random Amplification of Polymorphic DNA (RAPD) with a good discriminatory index of 0.99. Though a few clones were identified the majority were dissimilar. There was no specific major clone among our isolates.
E. coli and other genera of gram-negative bacteria possess a naturally occurring, chromosomally mediated β-lactamase and plasmid-mediated β-lactamase. Antibiotic misuse, ineffective empiric antibiotic therapy, poor dosing, prolonged antibiotic treatment and spontaneous use of these antibiotics in developing countries are the reasons for the development of high resistance rates in bacterial isolates.
There are a few lacunae in the present study. We did not test for NDM genes and CTXM. However
Conclusion
Phenotypic tests for beta-lactamase detection only confirm whether a beta-lactamase is produced but are unable to detect the beta-lactamase subtype and lead to failure of detection of those genes whose expression is hidden or masked. From our study, we have seen that among beta-lactamases ESBL production is high among uropathogens. This could be overlooked by routine disk diffusion methods and lead to inappropriate use of antibiotics and treatment failure. Hence, routine ESBL testing for uropathogens along with antimicrobial susceptibility testing would be useful for all cases of UTI. Also, a combination of both phenotypic and genotypic methods is suggested as the method of choice for the detection of betalactamase- producing strains of enterobacteriaceae. Sequencing is sensitive and can identify newer beta-lactamase types. Where possible this should be included for epidemiological reasons and periodic evaluation is needed.Funding
No separate funding was available for this study. Funding provided by the institute for routine microbiological testing was used.Conflict of Interest
The authors declare no conflict of interest.Availability of Data and Material
All data and materials of the study are available whenever required.Authors’ Contributions
MS designed, conceptualized the study and analyzed the data. TS collected the data, performed the laboratory testing and compiled the results. MS and TS wrote the entire manuscript. Both authors read and approved the final manuscript.Ethical Statement
Institutional ethical clearance has been obtained to use the isolates from our repository.References
- Niranjan V, Malini A. Antimicrobial resistance pattern in Escherichia coli causing urinary tract infection among inpatients. Indian J Med Res. 2014;139(6):945-948.
[Google Scholar] [PubMed]
- Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13(5):269-284.
[Crossref] [Google Scholar] [PubMed]
- Ahmed SS, Shariq A, Alsalloom AA, Babikir IH, Alhomoud BN. Uropathogens and their antimicrobial resistance patterns: Relationship with urinary tract infections. Int J Health Sci. 2019;13(2):48-55.
[Google Scholar] [PubMed]
- Asadi S, Kargar M, Solhjoo K, Najafi A, Ghorbani-Dalini S. The association of virulence determinants of uropathogenic Escherichia coli with antibiotic resistance. Jundishapur J Microbiol. 2014;7(5):e9936.
[Crossref] [Google Scholar] [PubMed]
- Dehbanipour R, Rastaghi S, Sedighi M, Maleki N, Faghri J. High prevalence of multidrug-resistance uropathogenic Escherichia coli strains, Isfahan, Iran. J Nat Sci Biol Med. 2016;7(1):22-26.
[Crossref] [Google Scholar] [PubMed]
- Miles TD, McLaughlin W, Brown PD. Antimicrobial resistance of Escherichia coli isolates from broiler chickens and humans. BMC Vet Res. 2006;2:7.
[Crossref] [Google Scholar] [PubMed]
- Muhammad A, Khan SN, Ali N, Rehman MU, Ali I. Prevalence and antibiotic susceptibility pattern of uropathogens in outpatients at a tertiary care hospital. New Microbes New Infect. 2020;36:100716.
[Crossref] [Google Scholar] [PubMed]
- Zhou M, Wang Y, Liu C, Kudinha T, Liu X, Luo Y, et al. Comparison of five commonly used automated susceptibility testing methods for accuracy in the China Antimicrobial Resistance Surveillance System (CARSS) hospitals. Infect Drug Resist. 2018;11:1347-1358.
[Crossref] [Google Scholar] [PubMed]
- Lee K, Chong Y, Shin HB, Kim YA, Yong D, Yum JH. Modified hodge and EDTA-disk synergy tests to screen metallo-beta-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 2001;7(2):88-91.
[Crossref] [Google Scholar] [PubMed]
- Black JA, Moland ES, Thomson KS. AmpC disk test for detection of plasmid-mediated AmpC beta-lactamases in Enterobacteriaceae lacking chromosomal AmpC beta-lactamases. J Clin Microbiol. 2005;43(7):3110-3113.
[Crossref] [Google Scholar] [PubMed]
Author Info
Tonushyam Sonowal* and Shariff MCitation: Sonowal T, Shariff M (2025) Phenotypic and Molecular Characterization of Multidrug-Resistant Uropathogenic Escherichia coli. Appli Microbiol Open Access. 11:350.
Received: 26-Feb-2024, Manuscript No. AMOA-24-29792; Editor assigned: 29-Feb-2024, Pre QC No. AMOA-24-29792 (PQ); Reviewed: 14-Mar-2024, QC No. AMOA-24-29792; Revised: 03-Feb-2025, Manuscript No. AMOA-24-29792 (R); Published: 10-Feb-2025 , DOI: 10.35248/2471-9315.25.11.350
Copyright: © 2025 Sonowal T, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.