Chemotherapy: Open Access
Open Access
ISSN: 2167-7700
ISSN: 2167-7700
Research Article - (2023)Volume 11, Issue 4
Objective: Limited evidence suggests a vital role for SEC11A in carcinogenesis. Our study conducted an integrated analysis on the molecular features and clinical relevance of SEC11A across pan-cancer based on multi-omics data.
Materials and methods: SEC11A expression was analyzed in 33 types of tumor and normal specimens utilizing transcriptome profiles from The Cancer Genome Atlas-Genotype-Tissue Expression (TCGA-GTEx) project and protromic data from Clinical Proteomic Tumor Analysis Consortium (CPTAC) project. Copy number alterations and methylation of SEC11A were investigated across pan-cancer with cBioPortal. Clinical relevance and prognostic implications of SEC11A were evaluated in The Cancer Genome Atlas (TCGA) cohorts. Gene Set Enrichment Analysis (GSEA) of SEC11A was conducted in Head and Neck Squamous Cell Carcinoma (HNSCC) with cluster profiler package. Correlation of SEC11A with immune cell infiltrations was estimated with Estimating the Proportions of Immune and Cancer cells (EPIC), Tumor Immune Estimation Resource (TIMER), xCELL, Microenvironment Cell Populations-counter (MCP-counter), quanTIseq, Cell type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT) and Immune Cell Abundance Identifier (ImmuCellAI) algorithms. Inhibitory Concentration (IC50) values of anti-cancer agents were curated from Genomics of Drug Sensitivity in Cancer (GDSC) project and their correlations to SEC11A were calculated.
Results: We found the expression of SEC11A was upregulated in most of cancer types (24/33), while was downregulated in 3 types of cancers. Genetic amplification in SEC11A was widespread across pan-cancer. There were negative correlations of SEC11A Ribonucleic Acid (RNA) expression with methylation level. The upregulation of SEC11A indicated unfavorable prognosis in adrenocortical carcinoma and HNSCC. Moreover, SEC11A was closely involved in cell cycle, TP53 regulation and antigen processing. SEC11A was positively associated with tumor-associated macrophage and fibroblast but negatively associated with CD8+ T cell. The expression of SEC11A was positively correlated to drug resistance to anticancer drugs.
Conclusion: These findings suggested that SEC11A could act as a prognostic biomarker across pan-cancer. Upregulation of SEC11A was in relation to tumor immunosuppressive status. Collectively, our results offered a valuable resource that might guide mechanistic and therapeutic analysis of SEC11A across cancers.
Pan-cancer; SEC11A; Prognosis; Immunosuppression; Resistance; Tumor microenvironment
TCGA: The Cancer Genome Atlas; GTEx: Genotype-Tissue Expression; RNA-seq: RNA sequencing; TPM: Transcripts Per Million; CNAs: Copy-Number Alterations; OS: Overall Survival; DSS: Disease-Specific Survival; DFI: Disease-Free Interval; PFI: Progression-Free Interval; GSEA: Gene Set Enrichment Analysis; GO: Gene Ontology; EPIC: Estimating the Proportions of Immune and Cancer Cells; CIBERSORT: Cell type Identification by Estimating Relative Subsets of RNA Transcripts; TIMER 2: Tumor Immune Estimation Resource 2; ImmuCellAI: Immune Cell Abundance Identifier; GDSC: Genomics of Drug Sensitivity in Cancer; ACC: Adrenocortical Carcinoma; LAML: Acute Myeloid Leukemia; OV: Ovarian Serous Cystadenocarcinoma; BLCA: Bladder Cancer; BRCA: Breast Cancer; CHOL: Cholangiocarcinoma; COAD: Colon Adenocarcinoma; DLBC: Lymphoid Neoplasm Diffuse Large B-cell Lymphoma; ESCA: Esophageal Carcinoma; GBM: Glioblastoma Multiforme; HNSCC: Head and Neck Squamous Cell Carcinoma; KIRC: Kidney Renal Clear Cell Carcinoma; KIRP: Kidney Renal Papillary Cell Carcinoma; LGG: Brain Lower Grade Glioma; LIHC: Liver Hepatocellular Carcinoma; LUAD: Lung Adenocarcinoma; LUSC: Lung Squamous Cancer; PAAD: Pancreatic Adenocarcinoma; PRAD: Prostate Adenocarcinoma; READ: Rectum Adenocarcinoma; SKCM: Skin Cutaneous Melanoma; STAD: Stomach Adenocarcinoma; TGCT: Testicular Germ Cell Tumors; THCA: Thyroid Carcinoma; THYM: Thymoma; UCEC: Uterine Corpus Endometrial Carcinoma; UCS: Uterine Carcinosarcoma; CESC: Carcinoma and Endocervical Adenocarcinoma; KICH: Kidney Chromophobe; MESO: Mesothelioma; PCPG: Pheochromocytoma and Paraganglioma; SARC: Sarcoma; UVM: Uveal Melanoma; Tgd: Gamma Delta T cell; Tfh: Follicular Helper T Cell; Tc: Cytotoxic T Cell; Tex: exhausted T; NKT: Natural Killer T Cell; Th 17: T helper cell 17 cell; nTreg: Natural Regulatory T; mRNA: messenger RNA; CPTAC: Clinical Proteomic Tumor Analysis Consortium; PPI: Protein-Protein Interaction; MCP-counter: Microenvironment Cell Populations-counter; GDSC 2: Genomics of Drug Sensitivity in Cancer 2; TGF-α: Transforming Growth Factor Alpha; EGFR: Epidermal Growth Factor Receptor; STRING: Search Tool for the Retrieval of Interacting Gevnes/Proteins
Cancer is a major burden globally and the leading cause of death. As estimated, there are 19.3 million new cancer cases and 10.0 million cancer deaths in 2020 [1]. This disease is a multi-step malignancy and featured by complex biological characteristics, involving multiple genetic and epigenetic alterations in the genome [2]. As high-throughput sequencing progresses, numerous studies have offered an in-depth survey of genetic dysregulation in cancers [3]. Analyses of molecular changes across pan-cancer identify commonalities and differences in critical biological processes that are dysregulated in tumor cells from distinct lineages [4].
Recent studies have proposed that SEC11A is a key regulator for carcinogenesis [5]. SEC11A is up-regulated in basal-like bladder cancer cases, which could be served as an independent prognostic predictor for bladder cancer [5]. Meanwhile, silencing SEC11A may weaken bladder cancer cell growth and invasiveness. SEC11A up-regulation is detected in gastric cancer and poorer survival in SEC11A-positive patients displays undesirable survival outcomes [6]. Forced expression of SEC11A may activate gastric cancer progression via promoting Transforming Growth Factor Alpha (TGF-α) secretion. Interestingly, study found that high SEC11A expression was examined in squamous cell carcinoma of tongue and its activation induced proliferation, migration and invasion in tongue squamous cell carcinoma cells [7]. SEC11A up-regulation is in relation to increased cancer-specific mortality among esophageal squamous cell carcinoma [8]. Moreover, SEC11A knockdown weakens cell proliferation as well as Epidermal Growth Factor Receptor (EGFR) signaling in esophageal squamous cell carcinoma. SEC11A exhibits up-regulated expression in parathyroid adenoma [9]. SEC11A-positive colorectal cancer patients usually display advanced pathological stage and SEC11A expression serves as an independent prognostic factor [10]. Collectively, SEC11A serves as an oncogene in cancer progression. Nevertheless, there is lack of pan-cancer evidence of the role of SEC11A on diverse cancer types on the basis of big clinical data. Growing evidence indicates that tumor microenvironment containing malignant, non-malignant, hematopoietic, as well as mesenchymal cells possesses clinic-pathological implications in prediction of therapeutic effects and survival outcomes of cancer patients [11]. Immune cells act as critical elements within the tumor stroma. Innate immune cells (macrophage, neutrophil, dendritic cell, innate lymphoid cell, myeloid-derived suppressor cell and natural killer cell) and adaptive immune cells (T cell and B cell) within tumor microenvironment participate in cancer development. The interplay between cancer cells and tumor microenvironment fosters tumor growth and metastases [12]. Oncogene-driven alterations in tumor cells may influence tumor microenvironment, thereby limiting immune response and presenting barriers to cancer therapy [13]. Hence, exploration of the association of SEC11A and tumor microenvironment may assist uncover the role of SEC11A in cancer progression.
Despite the presence of research on SEC11A gene, there is still a lack of researches about the expression, Copy Number Alterations (CNAs), methylation and mutations of SEC11A in cancers. In our study, we conducted a comprehensive analysis concerning identifying the molecular characteristics and clinical implications of SEC11A across diverse cancer types utilizing multi-omics data.
Retrieval of pan-cancer data
The Cancer Genome Atlas (TCGA), a web-based, freely accessible database, generates a large amount of next-generation sequencing data, including over 11,000 tumors in 33 cancer types. Genotype-Tissue Expression (GTEx) offers transcriptome profiles from 53 normal tissue sites across approximately 1,000 persons through publicly available RNA sequencing (RNA-seq). RNA-seq profiling of normal and primary tumors was retrieved from TCGA and GTEx via University of California Santa Cruz (UCSC) Xena repository (https://xena.ucsc.edu/). SEC11A messenger RNA (mRNA) expression in normal and tumor tissues across pan-cancer was visualized using ggplot-2 package [14]. Moreover, SEC11A mRNA expression in diverse tumor tissues or normal tissues was analyzed via ggradar package. Log-2[Transcripts Per Million (TPM)+1] transformed expression data were employed for the box plots.
Analysis of proteins interacted with SEC11A
Through the Clinical Proteomic Tumor Analysis Consortium (CPTAC); (https://proteomics.cancer.gov/programs/cptac), we curated the proteomic data of tumor and normal tissues across pan-cancer [15]. SEC11A protein expression was analyzed in tumor and normal tissues. Proteins interacted with SEC11A were analyzed through Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) [14]. The Protein-Protein Interaction (PPI) network was then constructed.
Genetic mutations and methylation analysis
Somatic Copy-Number Alterations (CNAs) and methylation levels of SEC11A were curated from the cBioPortal that is a repository of cancer genomics datasets [15]. Correlations of SEC11A expression with CNA values or methylation levels were determined across pan-cancer utilizing Pearson correlation analysis.
Survival analysis
Univariate-cox regression models were conducted to determine the associations of SEC11A with Overall Survival (OS), Disease-Specific Survival (DSS), Disease-Free Interval (DFI) as well as Progression-Free Interval (PFI) across pan-cancer in TCGA cohorts. OS was defined as the period between the date of treatment and date of death from any cause. DSS was defined as the period between the date of treatment and date of death from a specific disease. DFI was defined as the interval between treatment and disease recurrence or death. PFI was defined as the interval between treatment and disease worsening or death. The cutoff value of SEC11A was determined in each cell type with survminer and survival packages. Kaplan-Meier curves of OS were presented between high and low expression SEC11A groups. OS difference was estimated utilizing log-rank test.
Gene Set Enrichment Analysis (GSEA)
Pearson correlation analysis was utilized for screening the co-expressed genes of SEC11A with P<0.05 across Head and Neck Squamous Cell Carcinoma (HNSCC) specimens [16]. Afterwards, Gene Set Enrichment Analysis (GSEA) was carried out with cluster profiler package [17]. On the basis of Gene Ontology (GO) and reactome pathway databases, biological functions of SEC11A were investigated [18,19].
Immune infiltration analysis
Immunedeconv package offers an access to six algorithms for robust quantification of the abundance levels of immune cells based on bulk RNA-seq profiling of different cancer types, composed of Estimating the Proportions of Immune and Cancer cells (EPIC), Microenvironment Cell Populations-counter (MCP-counter), quanTIseq, Cell type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT), xCell as well as Tumor Immune Estimation Resource 2 (TIMER 2) [20-26]. Immune Cell Abundance Identifier (ImmuCellAI) database, a gene set signature-based algorithm, may precisely estimate the abundance of 24 immune cells covering 18 T-cell subpopulations based on gene expression profiling [27]. Herein, the abundance of immune cells was quantified by above algorithms across pan-cancer.
Drug response analysis
Genomics of Drug Sensitivity in Cancer (GDSC) project, a freely available public resource, covers drug sensitivity data in nearly 809 cancer cell lines and marker genes of responses to 192 anti-cancer drugs on the basis of 75,000 experiments [28]. Associations of SEC11A with IC50 values of anti-cancer drugs were estimated utilizing spearman correlation analyses. The top six drugs were visualized through ggplot-2 package.
Statistical analysis
Statistical analysis was achieved with R language. Comparison between two groups was conducted with student’s t test or Wilcoxon test. Meanwhile, comparison between multiple groups was analyzed with one-way variance analysis. Correlation analysis was carried out with Pearson or Spearman correlation tests. P<0.05 was indicative of statistical significance.
SEC11A mRNA expression across pan-cancer
Here, we analyzed the expression of SEC11A in 33 types of tumor and normal specimens from TCGA and GTEx projects (Figure 1A). Our results demonstrated that SEC11A was significantly down-regulated in Adrenocortical Carcinoma (ACC), Acute Myeloid Leukemia (LAML) and Ovarian Serous Cystadenocarcinoma (OV) tissues. Meanwhile, its expression was prominently increased in Bladder Cancer (BLCA), Breast Cancer (BRCA), Cholangiocarcinoma (CHOL), Colon Adenocarcinoma (COAD), lymphoid neoplasm Diffuse Large B-cell Lymphoma (DLBC), Esophageal Carcinoma (ESCA), Glioblastoma Multiforme (GBM), Head and Neck Squamous Cell Carcinoma (HNSCC), Kidney Renal Clear Cell Carcinoma (KIRC), Kidney Renal Papillary Cell Carcinoma (KIRP), brain Lower Grade Glioma (LGG), Liver Hepatocellular Carcinoma (LIHC), Lung Adenocarcinoma (LUAD), Lung Squamous Cancer (LUSC), Pancreatic Adenocarcinoma (PAAD), Prostate Adenocarcinoma (PRAD), Rectum Adenocarcinoma (READ), Skin Cutaneous Melanoma (SKCM), Stomach Adenocarcinoma (STAD), Testicular Germ Cell Tumors (TGCT), Thyroid Carcinoma (THCA), Thymoma (THYM), Uterine Corpus Endometrial Carcinoma (UCEC) and Uterine Carcinosarcoma (UCS). However, no significant difference in SEC11A expression was detected in cervical squamous cell Carcinoma and Endocervical Adenocarcinoma (CESC), Kidney Chromophobe (KICH), Mesothelioma (MESO), Pheochromocytoma and Paraganglioma (PCPG), Sarcoma (SARC) and Uveal Melanoma (UVM). Among different cancer tissues obtained from TCGA project, SEC11A displayed the highest mRNA expression in THCA and the lowest mRNA expression in ESCA (Figure 1B). Moreover, the expression of SEC11A was the highest in bone marrow and the lowest in brain among diverse normal tissues curated from GTEx project (Figure 1C).
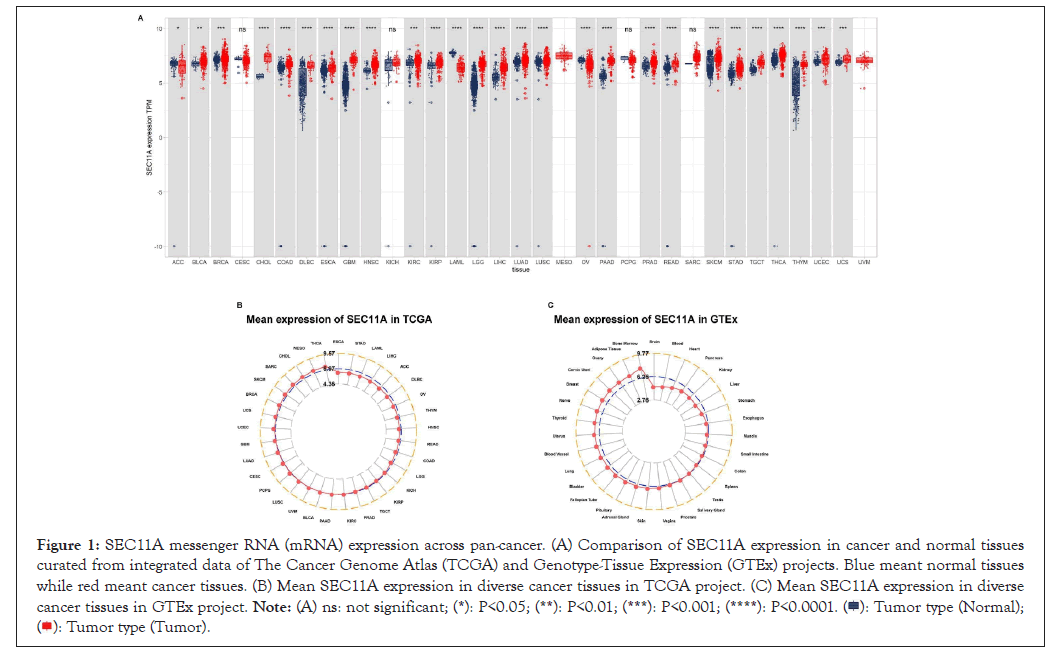
Figure 1: SEC11A messenger RNA (mRNA) expression across pan-cancer. (A) Comparison of SEC11A expression in cancer and normal tissues curated from integrated data of The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) projects. Blue meant normal tissues while red meant cancer tissues. (B) Mean SEC11A expression in diverse cancer tissues in TCGA project. (C) Mean SEC11A expression in diverse cancer tissues in GTEx project. 
Expression patterns of SEC11A mRNA in specific cancer types and pathological stages
We compared SEC11A expression in cancer and paired normal specimens. We observed the prominent up-regulation of SEC11A expression in BLCA, CHOL, ESCA, HNSCC, LIHC and STAD (Figures 2A-2F). In contrast, SEC11A was markedly down-regulated in KICH (Figure 2G). Moreover, this study presented the comparison of SEC11A expression in distinct pathological stages of each cancer type. Higher SEC11A expression was found in stage IV than stage III in BLCA (Figure 2H). Compared with stage II, SEC11A exhibited higher expression in stage IV in HNSCC (Figure 2I). In Figure 2J, we observed marked up-regulation of SEC11A in stage IV than stage I or stage III in KICH. In comparison to patient of stage I, higher SEC11A expression was observed in that of stage II and stage IV in LUAD (Figure 2K). As shown in Figure 2L, SEC11A displayed a significant up-regulation in stage IV than stage II for PAAD.
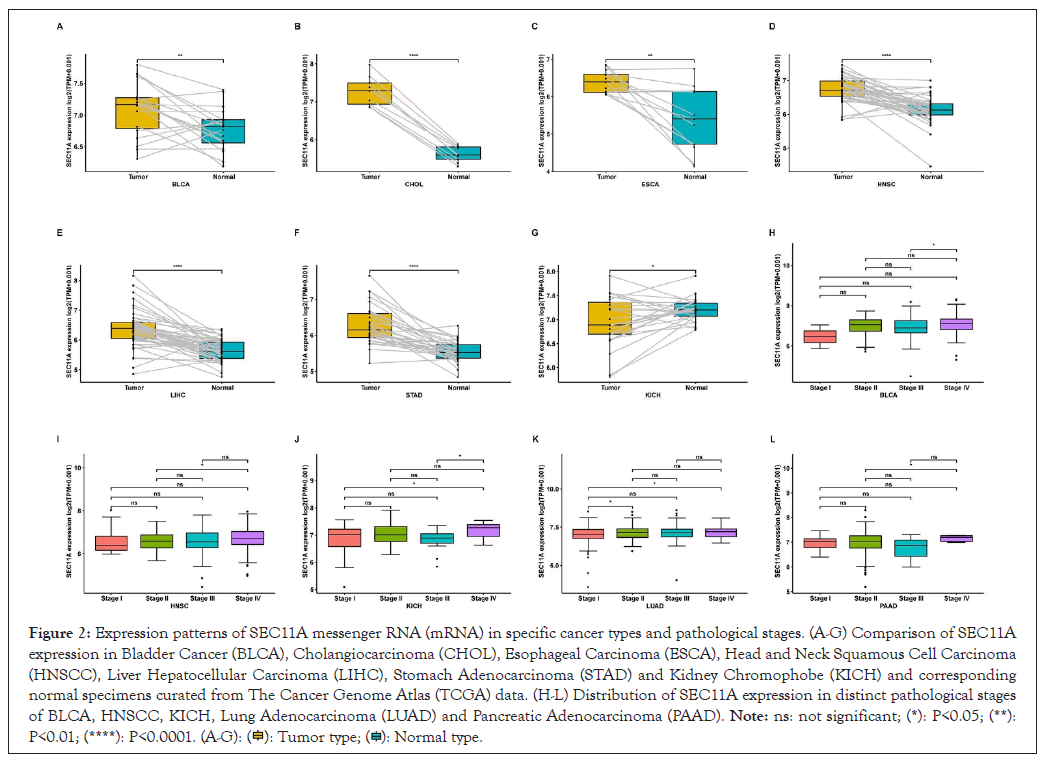
Figure 2: Expression patterns of SEC11A messenger RNA (mRNA) in specific cancer types and pathological stages. (A-G) Comparison of SEC11A expression in Bladder Cancer (BLCA), Cholangiocarcinoma (CHOL), Esophageal Carcinoma (ESCA), Head and Neck Squamous Cell Carcinoma (HNSCC), Liver Hepatocellular Carcinoma (LIHC), Stomach Adenocarcinoma (STAD) and Kidney Chromophobe (KICH) and corresponding normal specimens curated from The Cancer Genome Atlas (TCGA) data. (H-L) Distribution of SEC11A expression in distinct pathological stages of BLCA, HNSCC, KICH, Lung Adenocarcinoma (LUAD) and Pancreatic Adenocarcinoma (PAAD). 
Proteomic changes of SEC11A across pan-cancer and its interacted proteins
Through Clinical Proteomic Tumor Analysis Consortium (CPTAC) project, we evaluated the protein expression of SEC11A in tumor and normal tissues across pan-cancer. In Figure 3A, our results showed that SEC11A protein expression was markedly increased in BRCA, COAD, KIRC and UCEC compared with corresponding normal tissues. The PPI network revealed the potential proteins interacted with SEC11A, including SSR2, SEC61B, SEC11C, SPCS1, SPCS2, SPCS3, GCG, TMED10, GHRL and GIP (Figure 3B).
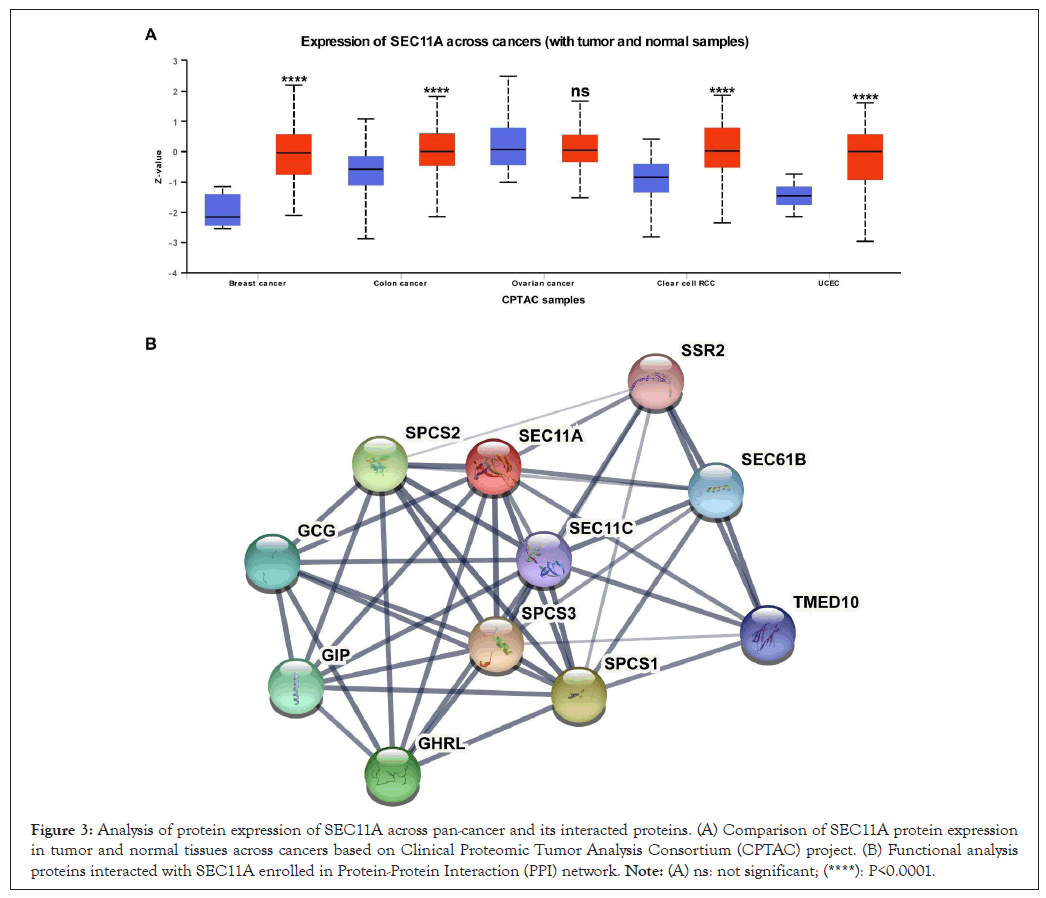
Figure 3: Analysis of protein expression of SEC11A across pan-cancer and its interacted proteins. (A) Comparison of SEC11A protein expression in tumor and normal tissues across cancers based on Clinical Proteomic Tumor Analysis Consortium (CPTAC) project. (B) Functional analysis proteins interacted with SEC11A enrolled in Protein-Protein Interaction (PPI) network. Note: (A) ns: not significant; (****): P<0.0001.
Genomic changes of SEC11A across distinct cancer tissues
Through cBioPortal, we observed genetic variations of SEC11A across distinct cancer tissues. In Figure 4A, amplification displayed the highest mutation frequency. This study further evaluated the association of SEC11A mRNA expression with linear copy-number value. Our data demonstrated that CNAs exhibited prominent correlations to SEC11A expression in most cancer types, except TGCT and KICH (Figure 4B). Moreover, we observed the prominently negative correlations of SEC11A expression with methylation levels across most of cancer types (Figure 4C). Hence, our findings indicated that high CNAs as well as low methylation contributed to high SEC11A expression across pan-cancer.
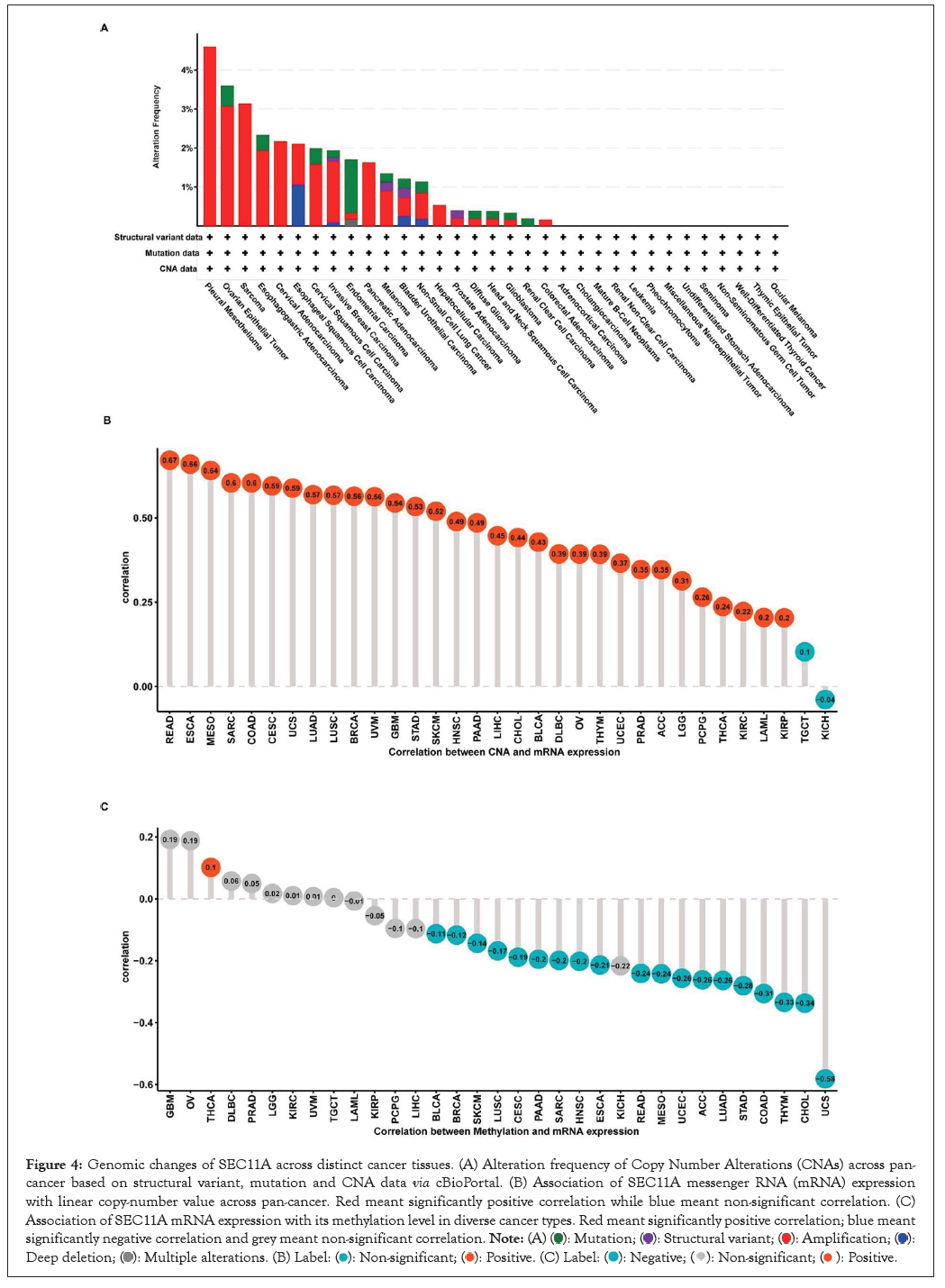
Figure 4: Genomic changes of SEC11A across distinct cancer tissues. (A) Alteration frequency of Copy Number Alterations (CNAs) across pan-cancer based on structural variant, mutation and CNA data via cBioPortal. (B) Association of SEC11A messenger RNA (mRNA) expression with linear copy-number value across pan-cancer. Red meant significantly positive correlation while blue meant non-significant correlation. (C) Association of SEC11A mRNA expression with its methylation level in diverse cancer types. Red meant significantly positive correlation; blue meant significantly negative correlation and grey meant non-significant correlation. 

Prognostic implication of SEC11A across pan-cancer
We assessed the implication of SEC11A in cancer prognosis curated from TCGA data. Univariate-cox regression models were conducted for investigating the association of SEC11A expression with OS, DSS, DFI and PFI across pan-cancer. In Figure 5A, SEC11A acted as a risk factor of OS of ACC and HNSCC as well as a protective factor of OS of KIRC and OV. In Figure 5B, SEC11A up-regulation indicated unfavorable DSS of ACC, HNSCC, LGG and PAAD as well as favorable DSS of OV. Also, we investigated that high SEC11A expression was in relation to unfavorable DFI for ACC, CESC, CHOL and KIRP patients (Figure 5C). As shown in Figure 5D, our data demonstrated that SEC11A up-regulation possessed the correlations to poorer PFI of ACC, HNSCC and LIHC as well as favorable PFI of KIRC and OV. Kaplan-Meier curves were then conducted for assessing the prognostic relevance of SEC11A across pan-cancer. Our data demonstrated that SEC11A acted as a risk factor of ACC, BLCA, CESC, COAD, HNSCC, KICH, LGG, LIHC, LUAD, MESO, PAAD, PDPG, PRAD, SARC, STAD and THYM (Figure 6).
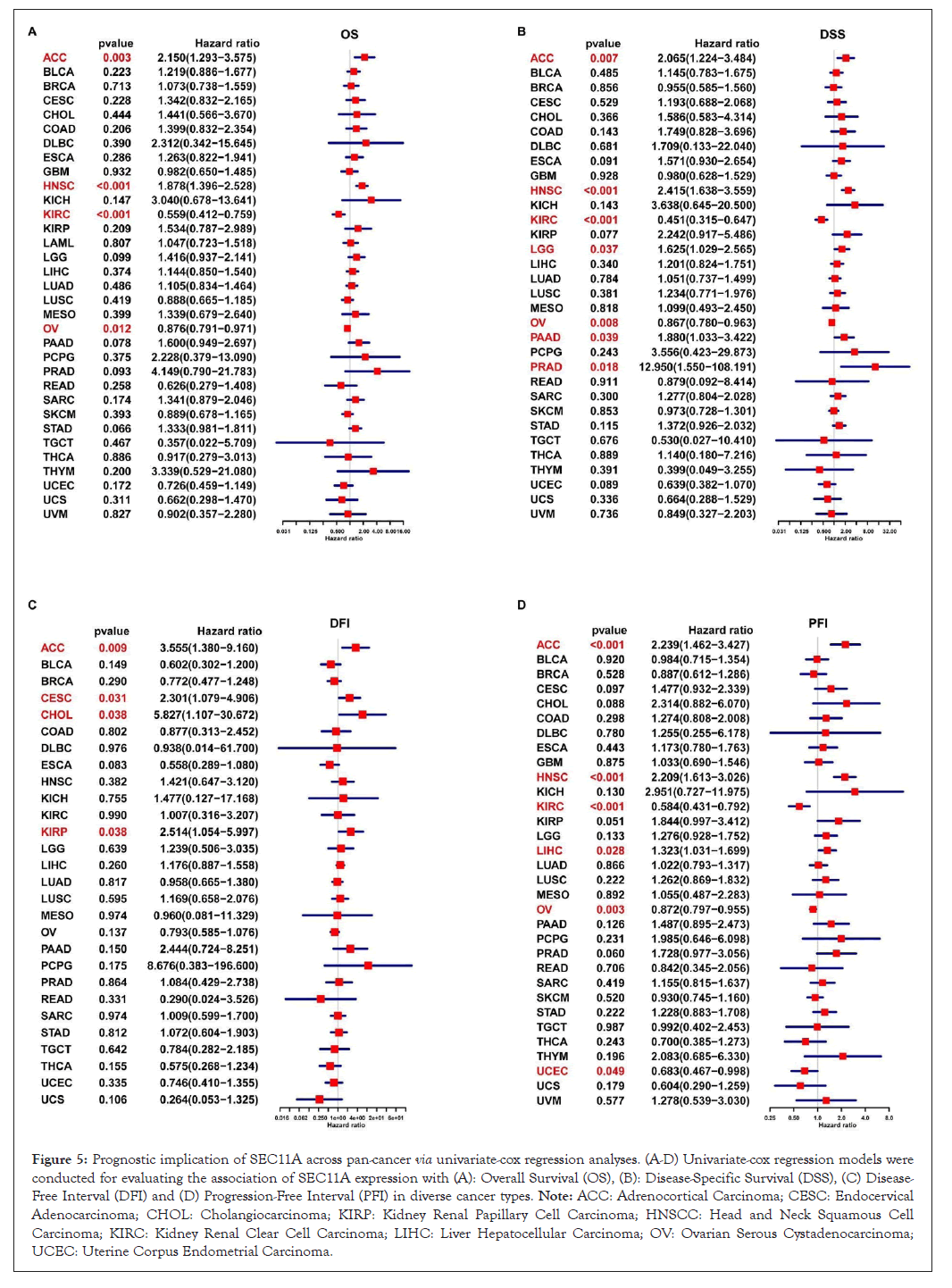
Figure 5: Prognostic implication of SEC11A across pan-cancer via univariate-cox regression analyses. (A-D) Univariate-cox regression models were conducted for evaluating the association of SEC11A expression with (A): Overall Survival (OS), (B): Disease-Specific Survival (DSS), (C) Disease-Free Interval (DFI) and (D) Progression-Free Interval (PFI) in diverse cancer types. Note: ACC: Adrenocortical Carcinoma; CESC: Endocervical Adenocarcinoma; CHOL: Cholangiocarcinoma; KIRP: Kidney Renal Papillary Cell Carcinoma; HNSCC: Head and Neck Squamous Cell Carcinoma; KIRC: Kidney Renal Clear Cell Carcinoma; LIHC: Liver Hepatocellular Carcinoma; OV: Ovarian Serous Cystadenocarcinoma; UCEC: Uterine Corpus Endometrial Carcinoma.
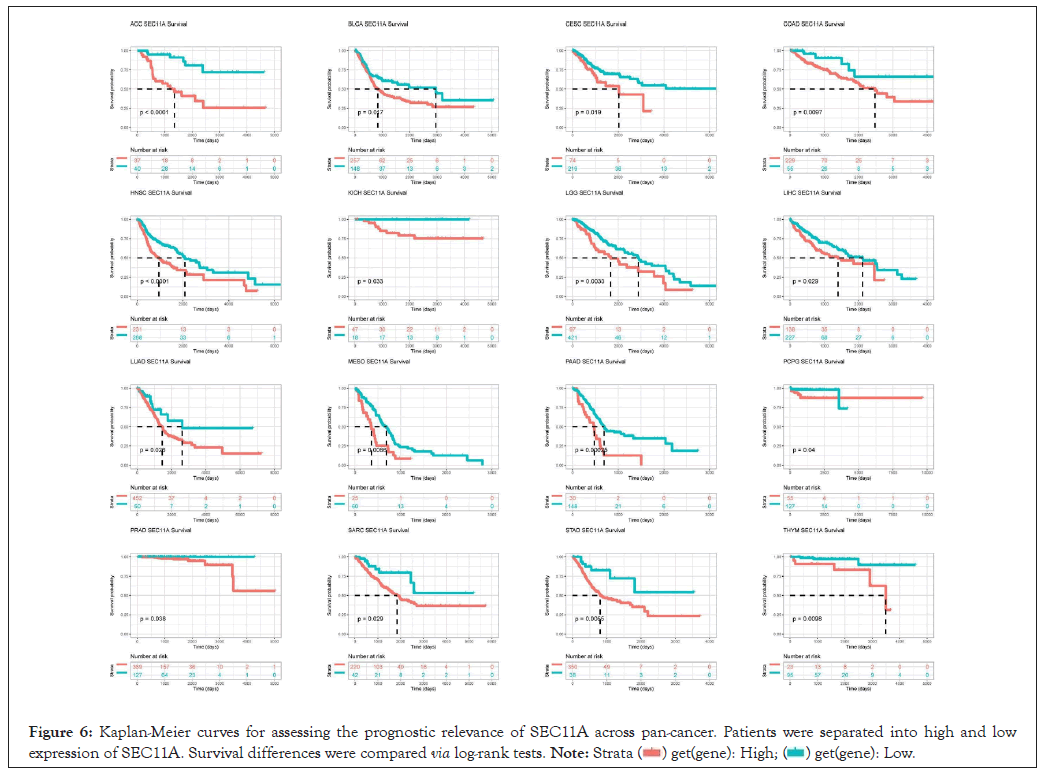
Figure 6: Kaplan-Meier curves for assessing the prognostic relevance of SEC11A across pan-cancer. Patients were separated into high and low expression of SEC11A. Survival differences were compared via log-rank tests. 
Prognostic implication of SEC11A in HNSCC subtypes
We also assessed the prognostic relevance in HNSCC subtypes. Our results demonstrated that high SEC11A expression was indicative of undesirable OS for HNSCC subtypes, including alveolar ridge, base of tongue, buccal mucosa, floor of mouth, hard palate, hypopharynx, larynx, lip, oral cavity, oral tongue, oropharynx, as well as tonsil (Figure 7).
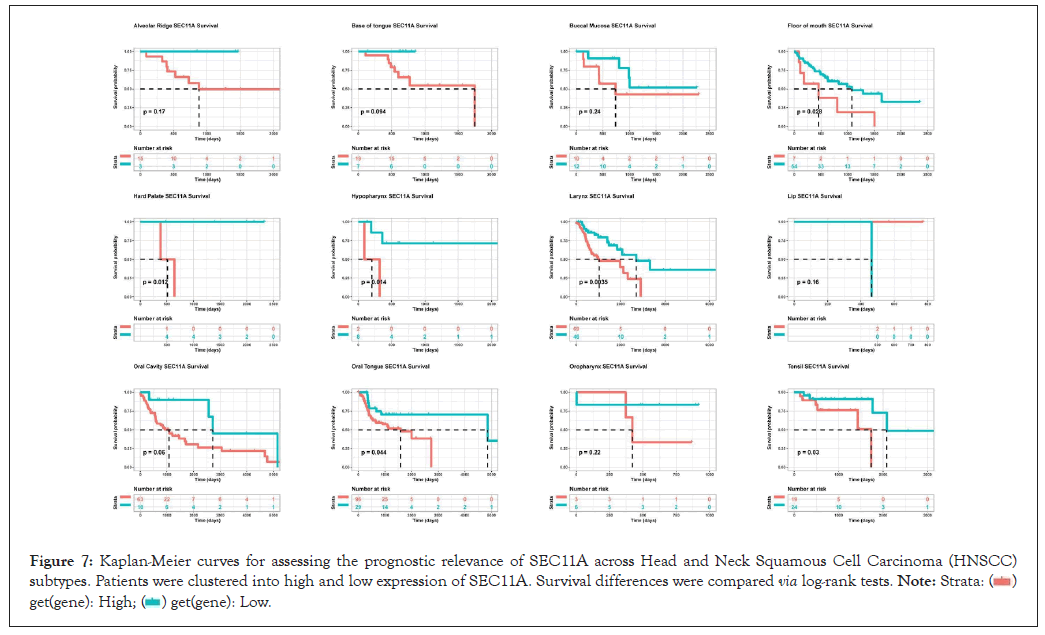
Figure 7: Kaplan-Meier curves for assessing the prognostic relevance of SEC11A across Head and Neck Squamous Cell Carcinoma (HNSCC) subtypes. Patients were clustered into high and low expression of SEC11A. Survival differences were compared via log-rank tests. 

Biological functions of SEC11A
To investigate the biological functions of SEC11A, GSEA was performed based on co-expressed genes of SEC11A that were identified via Pearson correlation analysis across HNSCC. The top 50 genes that were positively or negatively associated with SEC11A were separately shown in Figures 8A,8B. Based on GO database, we investigated that SEC11A was markedly correlated to RNA metabolism such as RNA splicing (Figure 8C). Based on the reactome pathway database, SEC11A displayed strong correlations with hallmarks of cancer such as cell cycle progression and TP53 activity as well as antigen processing (Figure 8D).
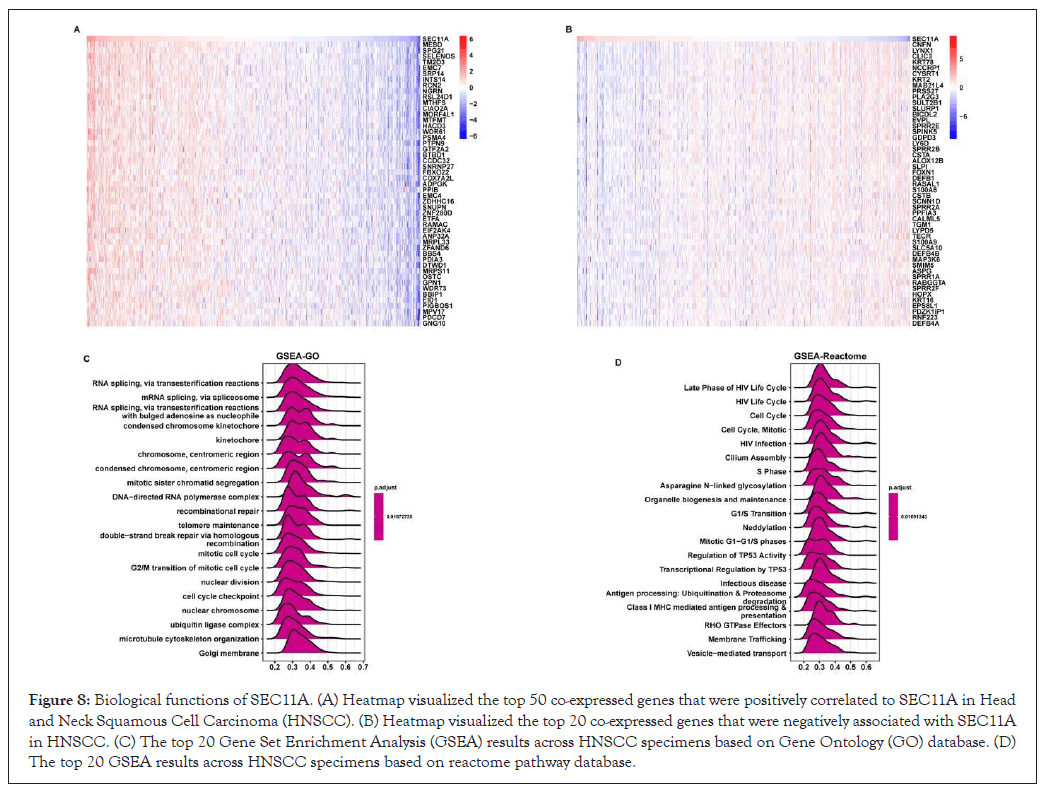
Figure 8: Biological functions of SEC11A. (A) Heatmap visualized the top 50 co-expressed genes that were positively correlated to SEC11A in Head and Neck Squamous Cell Carcinoma (HNSCC). (B) Heatmap visualized the top 20 co-expressed genes that were negatively associated with SEC11A in HNSCC. (C) The top 20 Gene Set Enrichment Analysis (GSEA) results across HNSCC specimens based on Gene Ontology (GO) database. (D) The top 20 GSEA results across HNSCC specimens based on reactome pathway database.
Association between SEC11A and immunosuppression
Utilizing EPIC, TIMER, xCELL, MCP-counter, quanTIseq and CIBERSORT algorithms, we estimated the abundance of tumor-associated macrophage and fibroblast and CD8+ T cell across pan-cancer tissues. We observed that SEC11A expression displayed positive correlations to tumor-associated macrophage and fibroblast while exhibited negative correlations to CD8+ T cell, especially HNSCC (Figures 9A-9C). Moreover, ImmuCellAI algorithm was applied for quantifying the abundance of immune cells especially including T cell subsets. In Figure 10A, SEC11A was negatively associated with CD8+ T cell, Natural Killer (NK) cell, gamma delta T cell (Tgd), B cell, follicular helper T cell (Tfh), cytotoxic T (Tc) cell, exhausted T (Tex) cell, CD4+ T cell and Natural Killer T (NKT) cell across pan-cancer. In contrast, SEC11A displayed positive correlations to T helper cell 17 (Th17) cell, monocyte, natural regulatory T (nTreg) cell and neutrophil across pan-cancer. These data suggested that SEC11A was in relation to immunosuppression. Especially, SEC11A displayed negative associations with immune infiltration score (r=-0.12, P=0.0074), NK cell (r=-0.19, P<0.0001) and CD8+ T cell (r=-0.33, P<0.0001) while had positive correlation to macrophage (r=0.12, P=0.0056) in HNSCC (Figures 10B-10E).
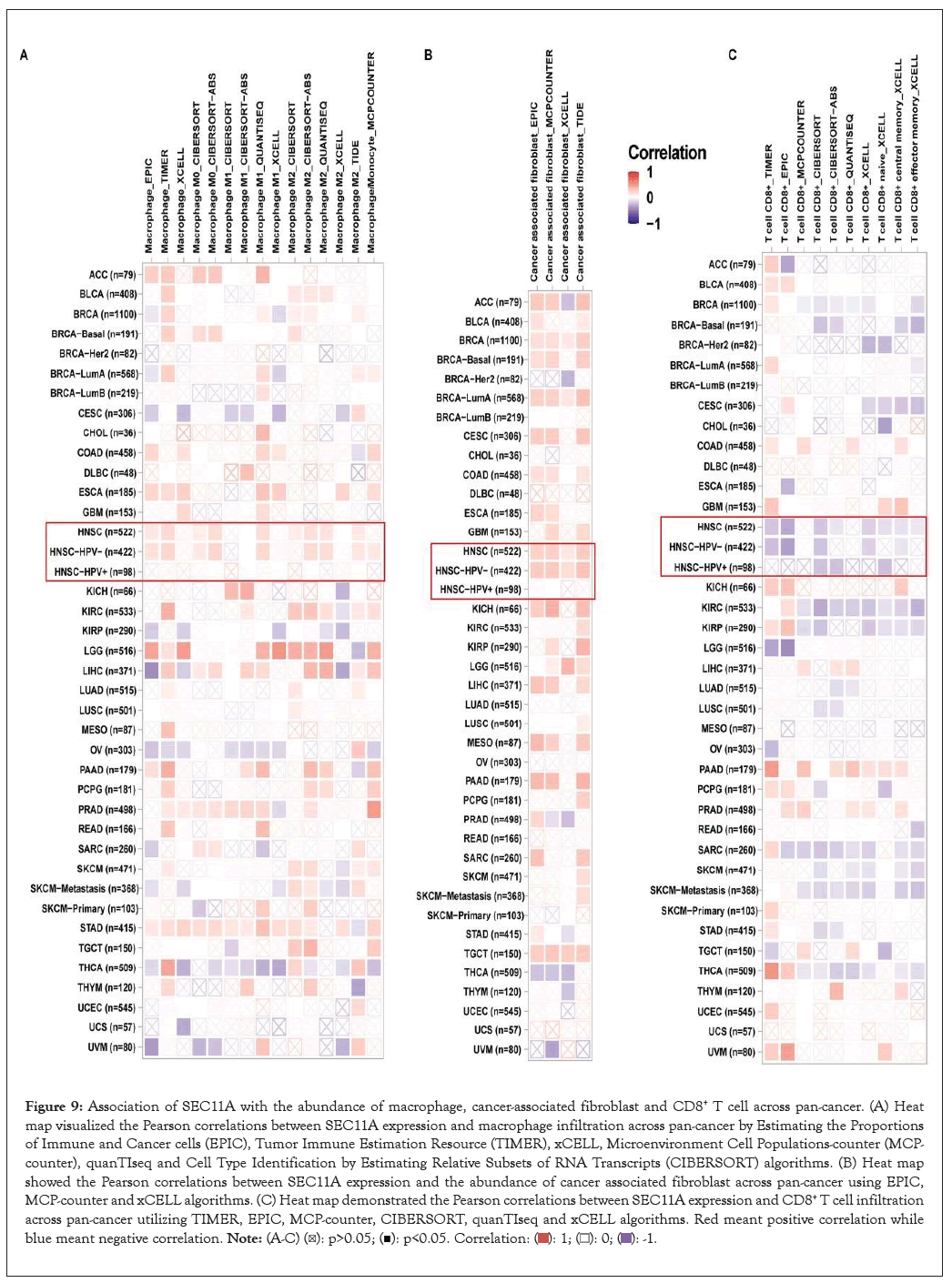
Figure 9: Association of SEC11A with the abundance of macrophage, cancer-associated fibroblast and CD8+ T cell across pan-cancer. (A) Heat map visualized the Pearson correlations between SEC11A expression and macrophage infiltration across pan-cancer by Estimating the Proportions of Immune and Cancer cells (EPIC), Tumor Immune Estimation Resource (TIMER), xCELL, Microenvironment Cell Populations-counter (MCP-counter), quanTIseq and Cell Type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT) algorithms. (B) Heat map showed the Pearson correlations between SEC11A expression and the abundance of cancer associated fibroblast across pan-cancer using EPIC, MCP-counter and xCELL algorithms. (C) Heat map demonstrated the Pearson correlations between SEC11A expression and CD8+ T cell infiltration across pan-cancer utilizing TIMER, EPIC, MCP-counter, CIBERSORT, quanTIseq and xCELL algorithms. Red meant positive correlation while blue meant negative correlation. 
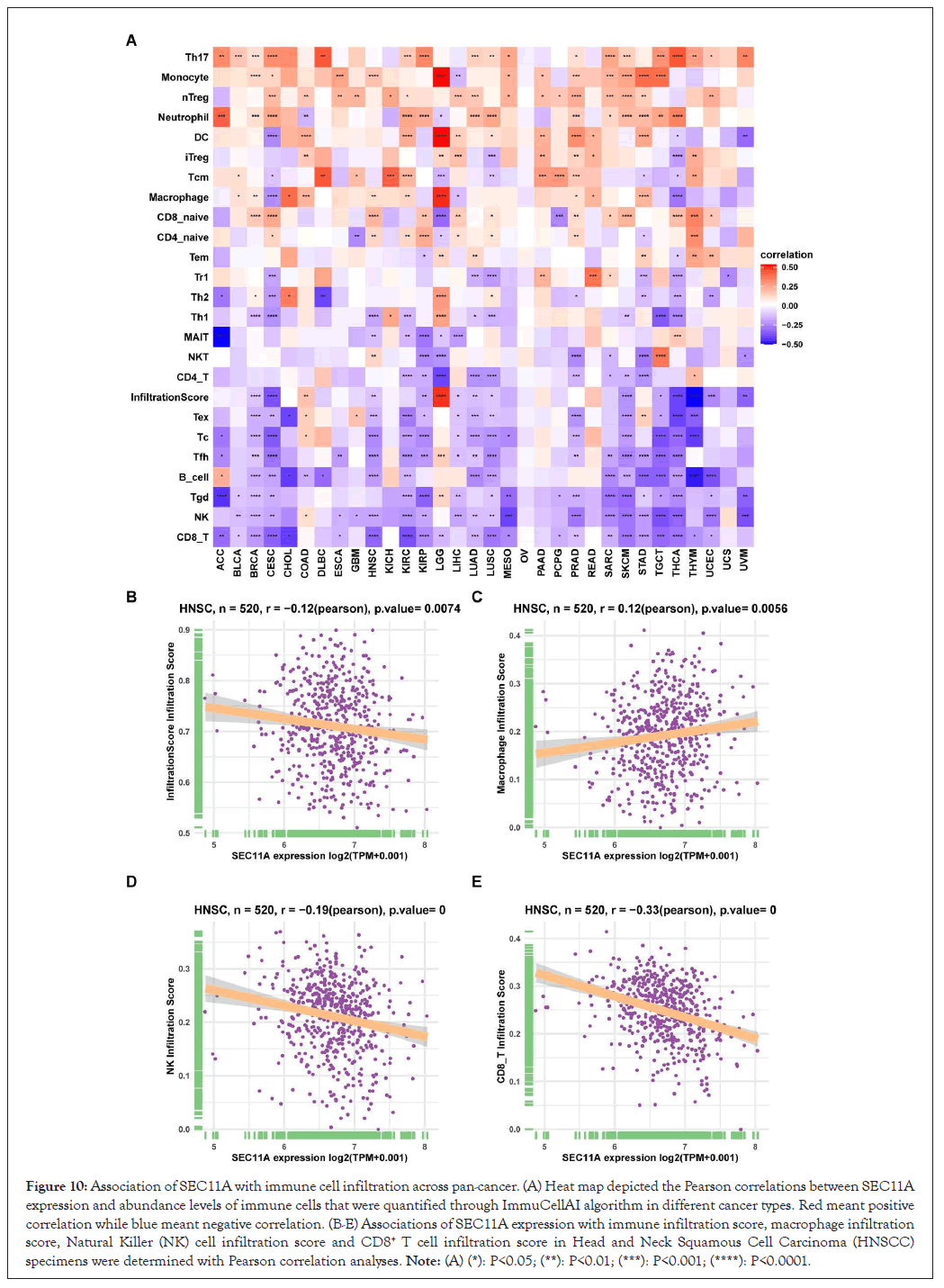
Figure 10: Association of SEC11A with immune cell infiltration across pan-cancer. (A) Heat map depicted the Pearson correlations between SEC11A expression and abundance levels of immune cells that were quantified through ImmuCellAI algorithm in different cancer types. Red meant positive correlation while blue meant negative correlation. (B-E) Associations of SEC11A expression with immune infiltration score, macrophage infiltration score, Natural Killer (NK) cell infiltration score and CD8+ T cell infiltration score in Head and Neck Squamous Cell Carcinoma (HNSCC) specimens were determined with Pearson correlation analyses. Note: (A) (*): P<0.05; (**): P<0.01; (***): P<0.001; (****): P<0.0001.
Association between SEC11A and drug resistance
Through GDSC project, we estimated the correlations of SEC11A expression with IC50 values of antitumor drugs utilizing spearman correlation analyses. The results were listed in Supplementary Table 1. Figure 11 visualized the top six antitumor drugs positively correlated with SEC11A expression, as follows: OSI-027 (r=0.23, P=1e-04), ABT737 (r=0.23, P<0.0001), AMG-319 (r=0.21, P<0.0001), MIRA-1 (r=0.21, P<0.0001), WEHI-539 (r=0.21, P<0.0001) and Entinostat (r=0.2, P<0.0001). The data above indicated that SEC11A exhibited positive correlations to resistance to these antitumor drugs.
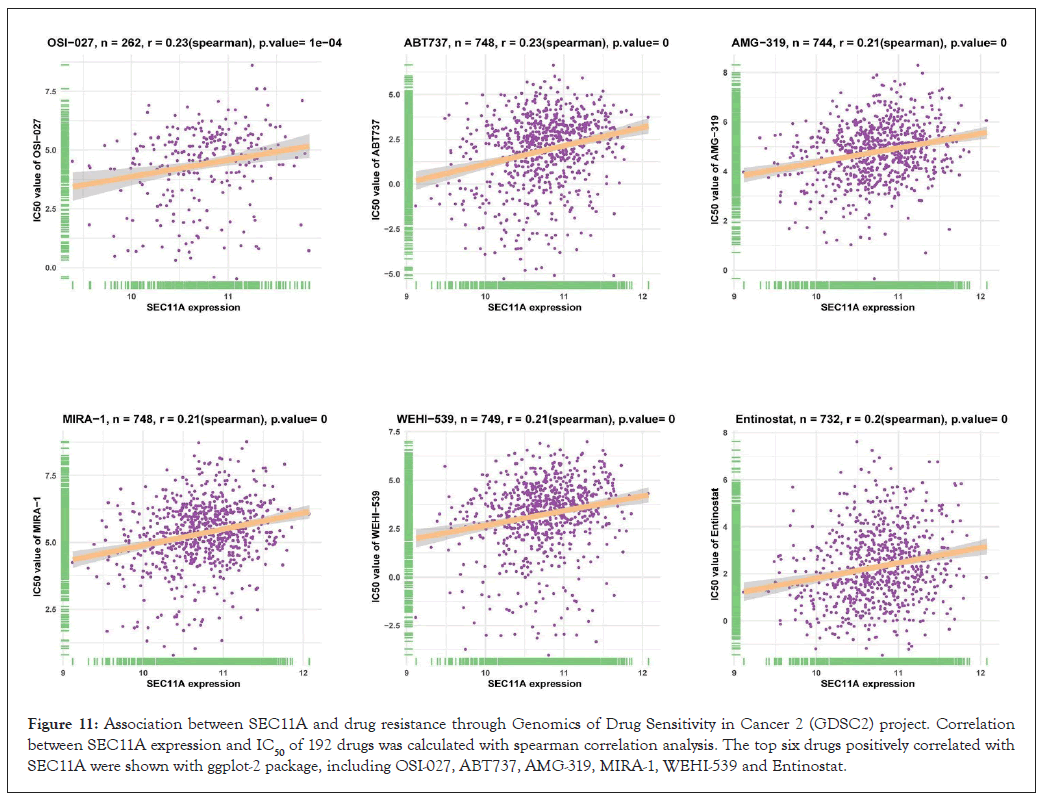
Figure 11: Association between SEC11A and drug resistance through Genomics of Drug Sensitivity in Cancer 2 (GDSC2) project. Correlation between SEC11A expression and IC50 of 192 drugs was calculated with spearman correlation analysis. The top six drugs positively correlated with SEC11A were shown with ggplot-2 package, including OSI-027, ABT737, AMG-319, MIRA-1, WEHI-539 and Entinostat.
In this study, we conducted comprehensive multi-omics analysis of SEC11A at transcriptomic, proteomic and epigenetic levels to observe the function of SEC11A across pan-cancer with an extensive manner. Multi-omics data can be utilized for predicting new functional interactions between molecules at diverse levels. Moreover, these data possess the potential for uncovering important biological investigations into hallmarks and pathways in comparison to single-omics analysis. This study carried out an integrated analysis of SEC11A, which aimed to offer an available resource concerning future related studies.
At the transcriptome and proteome levels, we observed the prominent up-regulation of SEC11A in most of cancer types. The Protein-Protein Interaction (PPI) network revealed the potential interacted proteins of SEC11A, including SSR2, SEC61B, SEC11C, SPCS1, SPCS2, SPCS3, GCG, TMED10, GHRL and GIP. CNAs are almost ubiquitous across pan-cancer as well as pose huge effects on tumor genome as an important type of genomic mutations [29]. Genetic CNAs exhibit the diverse landscape across pan-cancer [30]. Moreover, specific CNAs are in relation to cancer prognosis [31]. Our results demonstrated that SEC11A displayed the characteristics of increased copy number gain frequency across diverse cancer types. DNA methylation represents a common epigenetic mechanism, which participates in cancer progression [32]. This study investigated the methylation level of SEC11A across 33 cancer types. The methylation status of SEC11A was negatively correlated to SEC11A mRNA expression in most of cancer types, which demonstrated that there was homogeneity of SEC11A across cancers. DNA methylation in the gene body may enhance gene expression, while promoter methylation displays negative association with gene expression [33,34]. The negative association of SEC11A mRNA expression with methylation levels uncovered that methylation inhibitors might be applied on the basis of this promising therapeutic target SEC11A.
SEC11A displayed prominent correlations to several critical cancer hallmarks like cell cycle, regulation of TP53 activity, transcriptional regulation by TP53 [35]. Moreover, SEC11A was closely correlated with antigen processing. Neo-antigen plays a crucial role in cancer immunotherapy. SEC11A may induce a heightened neo-antigen-reactive T cell [36]. Our study might offer several clues to subsequent research concerning SEC11A. Tumor immune escape is an important step during cancer development, which has been the main cause about failure of several cancer immunotherapies [37]. Immunosuppressive cells promote tumor immune escape through inhibition of anti-cancer immune response [38]. Many translational research indicated that blockade of immunosuppressive cells or elimination of immunosuppression mechanism modulated by immunosuppressive cells or tumor cells may block tumor immune escape. Moreover, several clinical trials suggest that targeting immunosuppressive cells have achieved favorable clinical outcomes. T-cell subpopulations may contribute to long-term clinical benefits of antitumor therapy. Investigations of the abundance of immune cells may assist to have an in-depth understanding of the immune status as well as benefit cancer therapy. In this study, we employed different algorithms including MCP-counter, quanTIseq, CIBERSORT, TIMER-2 and ImmuCellAI to estimate the abundance of immune cells, particularly T-cell subpopulations based on transcriptome profiles. In this study, we observed the positive interactions between SEC11A and immunosuppressive cells in the tumor microenvironment.
OSI-027, a potent and selective dual inhibitor of Mammalian Target Of Rapamycin Complex 1/2 (mTORC1/2), possesses the anti-tumor properties such as cancers of the skin, breast, lung, as well as cervix [39]. B-cell lymphoma 2 (BCL2)/ BCL Extra-Large (BCLXL) inhibitor ABT737, is less effective against cancers because of resistance [40]. Small molecule MIRA-1 may induce anti-myeloma activity as well as synergize with anti-myeloma agents [41-43].
In this study, SEC11A mRNA expression exhibited positive correlations to drug resistance to most therapeutic agents. Future research is warranted to develop SEC11A inhibitors to alleviate drug resistance in cancer treatment. There are several possible limitations in our study. Some data are harvested from retrospective research where several clinical factors are not recorded as well as significant biases might affect choice of controls. Hence, more prospective clinical research is required for validating our findings.
Collectively, this study systematically investigated genetic and epigenetic changes and biological and clinical implications of SEC11A gene across pan-cancer, helping to further uncover the dysregulation of SEC11A in cancer. Our findings may be highly promising for personalized therapeutic regimens as well as clinical management.
Not applicable.
All of the authors approved the final version and publication.
Not applicable.
The authors have no conflict of interest to declare.
This study was supported by the National Natural Science Foundation of China (82002882).
Tingru Shao and Chengdong Liu: The initial conception and design of the manuscript, data curation, methodology, investigation, software, validation. Qingqing Wu and Xinyan Lu: Formal analysis, writing the original draft. Xiaozhi Lv and Tingru Shao: Funding acquisition, project administration, supervision, writing review and editing. All of the authors contributed towards drafting the manuscript and approved the final version.
We thank WeChat public account “Shengxinxiaoketang” for providing technical advice on our bioinformatics analysis.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Wu Q, Liu C, Huang G, Lu X, Lv X, Shao T (2023) Pan-Cancer Analysis Identifies SEC11A as an Immunological and Prognostic Biomarker. Chemo Open Access. 11:199.
Received: 28-Nov-2023, Manuscript No. CMT-23-28562; Editor assigned: 01-Dec-2023, Pre QC No. CMT-23-28562 (PQ); Reviewed: 15-Dec-2023, QC No. CMT-23-28562; Revised: 22-Dec-2023, Manuscript No. CMT-23-28562 (R); Published: 29-Dec-2023 , DOI: 10.35248/2167-7700.23.11.199
Copyright: © 2023 Wu Q, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.