Journal of Antivirals & Antiretrovirals
Open Access
ISSN: 1948-5964
ISSN: 1948-5964
Review Article - (2019)Volume 11, Issue 4
Dengue is a globally emerging health concern, still in preliminary stage of drug development. Many research scientists explored the dengue virus and identified its several targets that are categorized as structural and non-structural proteins, irrespective of its serotypic classification. A number of natural, synthetic and patented analogs are screened and studied against different serotypes and targets of dengue. Our review compiles the recent developments based on the huge number of such molecular space created by diverse scaffolds, specifically targeting NS2B-NS3 protease and methyltransferase (NS5) of dengue virus (DENV) infection. The emphasis of our article is to confer the leads towards the DENV drug discovery.
NS2B-NS3; NS5; Natural scaffolds; Synthetic scaffolds
DENV: Dengue Virus; RNA: Ribonucleic Acid; DF: Dengue Fever; DHF: Dengue Haemorrhagic Fever; WHO: World Health Organization; ADE: Antibody-Dependent Enhancement; ER: Endoplasmic Reticulum; NTPase: Nucleoside Triphosphatase; RTPase: RNA 5' Triphosphatase; MTase: Methyltransferase; RdRp: RNA-dependent RNA polymerase; mRNA: Messenger Ribonucleic Acid; SAM: S-adenosyl L-methionine; SAH: Sadenosyl L-homocysteine; EC50: Effective Concentration; CC50: Cytotoxic Concentration; IC50: Inhibitory Concentration; SI: Selective Index; μM: Micromole; μg: Microgram; nM: Nanomole.
Dengue virus (DENV), is a single stranded positive RNA virus encodes 10,173 nucleotides which are translated to form polyprotein [1], belonging to the genus Flavivirus and family Flaviviridae. DENV is arbovirus (arthropod-borne) and the vector responsible for transmission of infection is mosquitoes of Aedes genus, more specifically A. aegypti and A. albopictus [2]. DENV has four different antigenically distinct serotypes along with emerge of one more serotype recently viz DENV I, II, III, IV and V [3-5]. Among these serotypes, DENV II is most predominantly occurring virus. The main symptoms of infection are dengue fever (DF) and life threatening dengue haemorrhagic fever (DHF) causing drastic decrease in the blood platelet count [5].
Recently World Health Organization (WHO) announced with vaccine (CYD-TDV, or Dengvaxia®), is a live attenuated (recombinant) tetravalent vaccine, which is approved in seven countries (where major of the population affected from the disease) [2,6]. It is supposed to protect against all serotypes, but this task is difficult because of underlying mechanism of antibody-dependent enhancement (ADE) [7]. This necessitates the search for effective anti- dengue drug treatment. Tremendous research is going on to bring a novel drug molecule that can cater the need and evolve as potent against DENV. We have searched the available literature and chemical space from natural and synthetic sources in order to highlight evolution of novel molecule against DENV. Various naturally occurring plant phyto-constituents are proven to have anti-dengue property, which are discussed in this review article. The active molecules which are patented for their anti-dengue activity are also revealed in their respective sections.
Although there is lot of research is done in this area and still ongoing, but still till date formulated drug/medications is not available in the market to treat specifically DENV infection. The available treatment is still symptomatic. Hence, more systematic approach is needed to treat the DENV and eradicate it from the root, which is only possible by combining all the sources available with scientific literature.
DENV composed of a 5' untranslated region (UTR) attached with a cap, a single open reading frame (ORF), and a 3' UTR but it is devoid of 3' polyadenylate tail [8]. The main function of ORF is to encode long polyprotein, cleavage of these polyprotein by a host as well as viral proteases results into 10 viral proteins in which three are structural and seven are non- structural proteins. Numerous studies have been investigated using different approaches to target these structural and non-structural proteins, so that they can interfere with the entry of virus (targeting structural) or replication (targeting non-structural) to prevent DENV infection. The structural proteins attached to N-terminal consist of capsid, membrane and envelope whereas nonstructural proteases consist of NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5 is attached to C-terminal. The structural protein of DENV is shown in Figure 1.
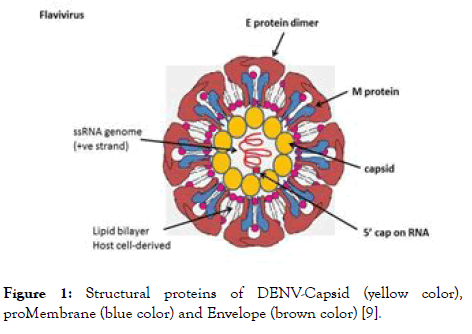
Figure 1. Structural proteins of DENV-Capsid (yellow color), proMembrane (blue color) and Envelope (brown color) [9].
Structural proteins of DENV
The structural proteins of DENV are projected as important targets for entry and fusion inhibitors.
Capsid (C): The structural proteins of DENV are projected as important targets for entry and fusion inhibitors. The capsid protein which forms the protective layer surrounding the genome of DENV contains hydrophobic C terminal region, central portion and highly basic region [10]. The C terminal contains a signal sequence which plays a dual role of anchoring the protein into the endoplasmic reticulum (ER) membrane and partitioning the proMembrane protein to the membrane [11]. The central portion of capsid promotes association of capsid protein with ER membrane [12]. This hydrophobic region of capsid could interact with the lipid molecules [13]. The highly basic region of capsid protein is associated with the interaction with the viral genome [14,15]. These all together makes the viral assembly more effective for further process [16]. When the virion recognizes the host receptor, it initiates fusion of the virion with endosome. This is followed by endocytosis and the capsid uncoats to release the viral genome into the cytoplasm for further transcription and translation purpose. Consequently, the capsid is necessary for packaging and release of mature viral particles from the cell [3,17].
proMembrane (prM): The prM is the precursor of the M protein, which provides resistance to low pH environment and protects the envelope protein from premature fusion during transit through the acidic environment of the trans-golgi network. This mechanism of protection is mainly due to the hydrophilic N-terminal portion of the protein which codes for the glycosylated “pr” segment of the prM protein [18]. Cleavage of this part from prM generates the M protein. This cleavage is mediated by furin, a protease and is required for virus maturation [19] and infection in cell culture [20].
Envelope (E): Envelope of DENV denotes as E protein which plays crucial role in recognizing the cellular receptor, fusing the viral assembly to cellular endosomic membrane. E protein belongs to class II fusion protein comprised of-central domain (domain I-N-terminal portion of the protein), a dimerization domain (domain II), and a receptor binding domain (domain III), which shown to be involved with antibody neutralization [21-25]. L-SIGN, GRP78, DC-SIGN, the high affinity laminin receptor and the mannose receptor -these are the different receptors which are involved in binding to E [2]. During process of binding of virus to the host cell, structural rearrangement occurs due to acidic environment of the enzyme which induces the fusion of cellular and viral membranes [26]. These rearrangements lead to the different changes in the conformations of E-protein [27].
Non-structural proteins of DENV as potential drug targets
The non-structural proteins consist of NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5, in which NS2B-NS3 protease complex- NS3 protease [helicase/nucleoside triphosphatase (NTPase)/RNA 5' triphosphatase (RTPase)] along with cofactor NS2B, and NS5 [Methyltransferase (MTase)/RNA-dependent RNA polymerase (RdRp)], plays a crucial role in virus replication. These two can serve as a potential target for antidengue agents since they are essential for virus replication. The role of NS1 is involved in RNA replication. It is a soluble protein detected very early during infection, but has received little attention so far as an antiviral target. NS2A is associated with the process of replication and is involved in assembly and interferon while NS2B is a cofactor for NS3. NS4A and NS4B are membrane-associated proteins believed to anchor and regulate the replication complex during the virus life-cycle and NS5 is a catalytic subunit of viral replication complex [28]. The graphical representation of non-structural proteins along with structural proteins is shown in Figure 2.
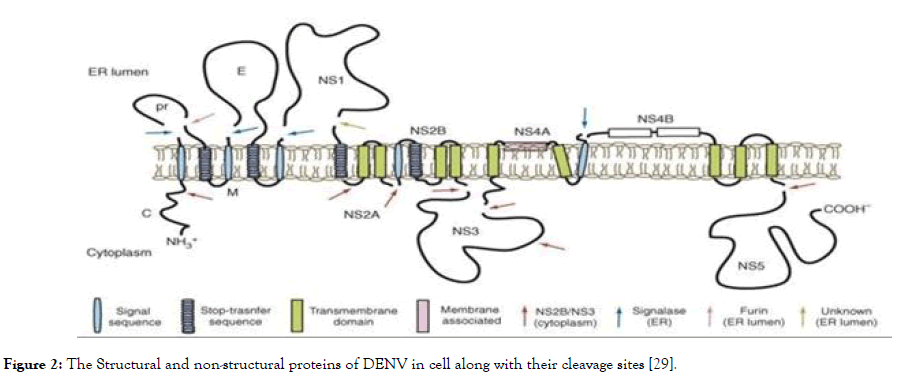
Figure 2. The Structural and non-structural proteins of DENV in cell along with their cleavage sites [29].
In this review we focus on two non-structural protein targets such as NS2B-NS3 protease complex and NS5 protease. We explored these two proteins based on the knowledge available on their 3D structure, their role in DENV replication and the reported literature about various molecules screened to inhibit these proteins. This review which discuss in detail about various scaffolds under study and expose the extensive chemical space to highlight the important structural features of all those evaluated scaffolds for the development of new molecules against DENV.
NS2B-NS3 complex: NS3 protein is of 618 amino acids and known as a multifunctional protein that functions both as a trypsin-like serine protease as well as RNA helicase and RNA triphosphotase (RTpase)/Nucleoside triphosphotase (NTpase). NS3 protein is made up of two important domains called as Nterminal protease domain (with 1-180 residues) and C-terminal helicase domain (with 180-618 residues). The enzyme consists of six β-strands that form two β-barrels with the conserved catalytic triad (His51, Asp75 and Ser135) sandwiched between them [30-32]. The DENV protease activity is due to the complex of NS2B-NS3 and the N-termini of several non-structural proteins are produced through cleavage by NS2B-NS3 at dibasic sites [33]. NS2B consist of hydrophilic core domain and hydrophobic domain. Studies showed that, 40 amino acids of hydrophilic sequence from the central region, is the required region in NS2B [34-36]. Further studies have confirmed that this region is sufficient to activate NS3 protease in vitro [36,37]. This sequence commonly called as hydrophilic core sequence or cofactor [38]. NS3 protein included with residues of this cofactor NS2B, and both are connected via a Gly-Ser linker to the N-terminus of full length NS3 [39]. Moreover, the NS2B wraps NS3 protease domain and forms belt-like structure-fundamental part of the NS2B-NS3 active site. This protease complex is anchored by NS2B, during the replication cycle in the ER membrane [36]. The N-terminal protease domain locates on upper portion of helicase domain. The protease domain is inter- connected with the linker to helicase domains and is also conserved region across all flaviviruses. It is acting as crucial for association of two domains in NS3 [40]. It is indicated that the basic surface residues of the protease domain are implicated in the binding of the helicase domain to its nucleic acid substrate [39-41].
The C-terminal helicase domain has seven structural motif resembles of super family 2 helicase [42]. The structure of helicase domain reveals to contain 3-subdomains viz I, II and III, with significant sequence identity and structural similarity to other flaviviruses helicase [43,44]. Sub domain I and II poised of six-standard β-sheet arranged in the central and surrounded with four α-helices with 181-326 and 327-481 residues respectively. These are similar kind of domains available with Hepatitis C virus and may act by similar mechanism [45-47]. Whereas Sub domain-III is a unique to flaviviruses, predicted to be a protein binding site. It contains 482- 618 residues and has three short α- helices which encircle the four parallel α-helices and two solvent exposed antiparallel β-strands [40].
The RTpase activity of NS3 involves in capping nascent viral RNA and whereas helicase along with NS2B manages the unwinding of double stranded replicative form of RNA [48]. However, the melting of secondary structures before initiation of RNA synthesis as well as for unwinding RNA duplexes chiefly controlled by the helicase and NTpase regions [44,48-50]. The helicase domain also interacts with the polymerase, NS5 [51].
NS5 protein: Methyltransferase (MTase) and polymerase
NS5 protein being a largest with 900 residues and most conserved in DENV, known to be involved in viral replication via catalytic subunit [6,52,53]. The mRNA of DENV possesses a type I cap structure (me7GpppA-me2) at its 5' end, which is essential for constancy and effectual translation during infection [54-57] and in this process NS5 protein plays a vital role. NS5 protein consists of N-terminal MTase domain (1-296 residues) and C-terminal RNA dependent- RNA polymerase (RdRp polymerase with 320-900 residues). The NS5 MTase takes globular fold- confirmed by X-ray crystallography and it is divided into N-terminal region (1-58 residues), a core domain (59-224 residues) and a C-terminal region (225-265 residues) [58,59]. The core domain has two specific binding sites Sadenosyl L-methionine (SAM, a cofactor and a methyl donor of NTase) and RNA binding site. This MTase domain sequentially catalyzes both guanine N-7 and ribose 2'-O-methylation [60,61]. The capping of mRNA involved with four steps in which the final two steps are catalyzed by MTase using SAM that generates SAH (S- adenosyl-L-homocysteine) as a byproduct [57,61,62]. Different research studies were conducted to reveal the importance of MTase and it is observed that the defect in N7 methylation eliminate the DENV replication which is totally controlled by MTase [54,63]. Hence NS5 MTase can serve as another putative target apart from NS2B-NS3 complex. Whereas the main concern while designing the molecules against DENV targeting MTase is selectivity; because of the existence of human MTase that is implicated in methylating protein, RNA and DNA [64]. The human MTase is devoid of specific pocket [65] in the vicinity of the SAM binding site which is present in Flaviviruses. Hence, by incorporating specific features in the molecules, which can help to occupy this pocket can inhibit specific SAM, that can result into decrease in the toxicity which is the resultant of non-specific inhibition of SAM. The structure of NS5- polymerase domain was observed and it is analogous to other RdRp molecules [66]. The catalytic mechanism involves incorporation of nucleotides coordinated by conserved aspartic acid residues (also known as GDD motif) with the help of two metal ions. The RdRp possesses nuclear localization signal (NLS) region which is stuck between 320-405 residues. The NLS region of RdRp is a structural component which is well defined [67-70]. The NS5 conserved residues 320-368 are involved in the interaction with NS3 [51,69]. RdRp also involves in the production of -ve sense RNA (-ss RNA) from the +ve sense viral RNA (+ss RNA) template which serves as a template for generating further more +ss RNA strands, these will be utilized either in protein translation or assembly to infectious virions [67,71,72]. RdRp contributes greater part in replication cycle like the replication of viral RNA and translation.
Replication cycle of DENV: The replication cycle of DENV is entirely pH dependent process in which dendritic cells and other cells of the immune system would serve as the primary host targets [73-75]. The entry of virus particles starts with the recognition of host cell receptors followed by fusion of virus via receptor mediated endocytosis, which is believed to utilize many receptors. At pH 6.0, the viral assembly gets disassembled in endosome; the capsid uncoats the genome i.e. single-stranded, +ss RNA and the genome gets released into the cytoplasm. This +ss RNA get translated via ribosomes to form viral polyprotein as well as get transcripted to form negative sense RNA with participation of replication complex to form replicates of +ss RNA, which is cleaved by host to yield structural and nonstructural viral proteins. This process and the replication occur at the surface of ER. At pH 7.2, the inclusion of viral proteins into ER and genome assembling and packaging of immature virus occurs followed by releasing of this immature virus by ER to reach trans-Golgi network at pH 6.7. The final maturation of virus is mainly due to decrease in the pH from 6.7 to 6.0, leading to the conformational changes in the structural proteins, as well as the proteolytic cleavage of prM by the host protease furin. The matured virus gets way out from the infected cells via exocytosis. As the whole replication process has been influenced by the microenvironment of the replication complex, it can be the major factor which can influence the various physical, chemical and cellular aspects of the targets.
The composition and replication cycle of DENV, indicate many target sites in both structural and non-structural proteins along with different stages of virus replication. Many researchers are motivated to investigate and are in the process of developing a therapeutic agent against dengue with more selectivity along with lesser side effects. With all the targets available, the nonstructural, NS2B-NS3 complex and NS5 evolved as putative targets which attracted many researchers. In this process, many natural scaffolds and synthetic scaffolds based on natural phytoconstituents are screened, studied and tested for their potency against all serotypes of DENV or with respect to any single serotype by targeting above proteins. Our review will portray several scaffolds that are targeted against these proteins. The scaffolds may be purely from natural origin, tested in the form of extracts or single constituent for anti-dengue property. The other scaffolds are with synthetic origin evaluated against dengue and few of them specifically reported as patented molecules for their activity against dengue.
Natural scaffolds targeting DENV
Nature emerged as a vast reservoir of phyto-constituents which can treat many diseases. Different herbal drugs containing flavonoids, quinones, alkaloids, polysaccharides and terpenoids are identified as anti-dengue agents. We further discuss these naturals according to their inhibitory activity on NS2B-NS3 and NS5.
The extract of Carica papaya, was well known for treating DENV infection. The studies indicated that the drastic reduction in the platelet count is the major symptom of dengue infection which is controlled by the extract. To avoid progression of the disease to severe form can be achieved by controlling the normal platelet count [76]. The main constituents of Carica papaya (Figure 3) are quercetin (1), caffeic acid (2), p-coumeric acid (3), kaempferol (4), protocatechic acid (5), chlorogenic acid (6), and 5,7- dimethoxy coumarin (7). The study involved use of bioinformatic tools to investigate the main constituent which is responsible for activity against dengue 2 virus (DENV-2). It is reported that the quercetin indicated highest binding energy with formation of six hydrogen bonds with the critical residues responsible for the activity of NS2B- NS3 protease. Caffeic acid showed four hydrogen bonding interactions and p-coumeric acid showed best binding affinity in terms of hydrogen bonding, but with poor ligand-receptor atom pair interaction. The remaining constituents formed five hydrogen bonding with the catalytic triad residues of NS2B-NS3 but compared to quercetin showed lower binding affinity [77]. The basic scaffold of quercetin contains fused benzo-dihydropyranone ring substituted with aromatic nucleus, resembling to that of the flavonoid scaffold.
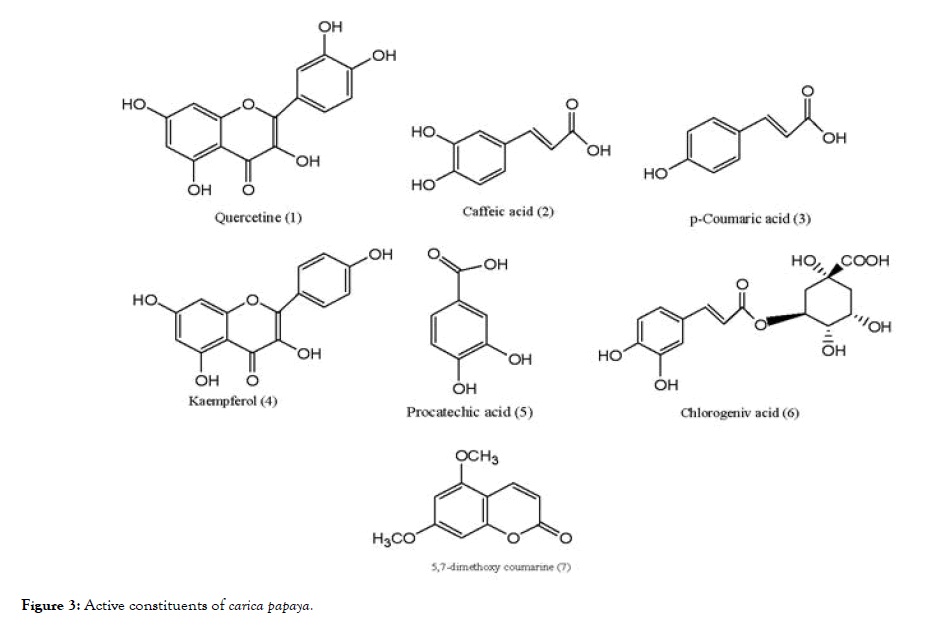
Figure 3. Active constituents of carica papaya.
Five Flavonoids from Mexican Tephrosia species were isolated and evaluated using LLC-MK2 cells by in vitro plaque assay method for DENV-2 serotype. Out of five flavonoids (8-12) shown in Figure 4, only glabranine (8) and 7-o-methylglabranine (9) showed significant inhibitory activity, rest of three are devoid of antiviral effect. Considering the structural features of compounds (8-12), it is apparent that structure activity relationship exists among flavonoids since glabranine (8) and 7- o-methylglabranine (9), which both contains a phenyl side-chain at C- 8 and the benzene ring is substituted with alkyl chain, were active as replication inhibitors [78]. In case of (10-12), although the basic scaffold is similar to that of (8) and (9), the open alkyl chain is cyclized, which might be the reason for their inactivity. The scaffold of these active flavonoids can be correlated with the quercetin, previously reported as active molecule.
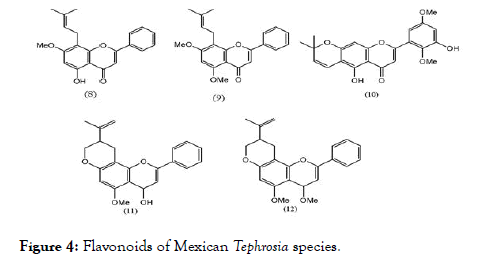
Figure 4. Flavonoids of Mexican Tephrosia species.
Another plant – Houttuyniacordata Thunb (Saururaceae; H. cordata) commonly known as “Pak Kan Thong” in Thai, tested against DENV-2, showed that the H. cordata extract (10-100 mg/mL) when incubated before and after the infection using HepG2 cells indicated reduction of intracellular DEN-2 RNA production that further link towards the reduction in the expression of dengue protein. The effective concentration (EC50) was 0.8 mg/mL. To determine the active phytoconstituents which are responsible for the activity Highperformance liquid chromatography (HPLC) analysis were conducted and it was found that hyperoside (13) (Figure 5) was the predominant bioactive compound, and which might be responsible for inhibition [79]. The structure of hyperoside, similar to previously reported flavonoid molecules along with substitution of additional glycoside, which might have made it active.
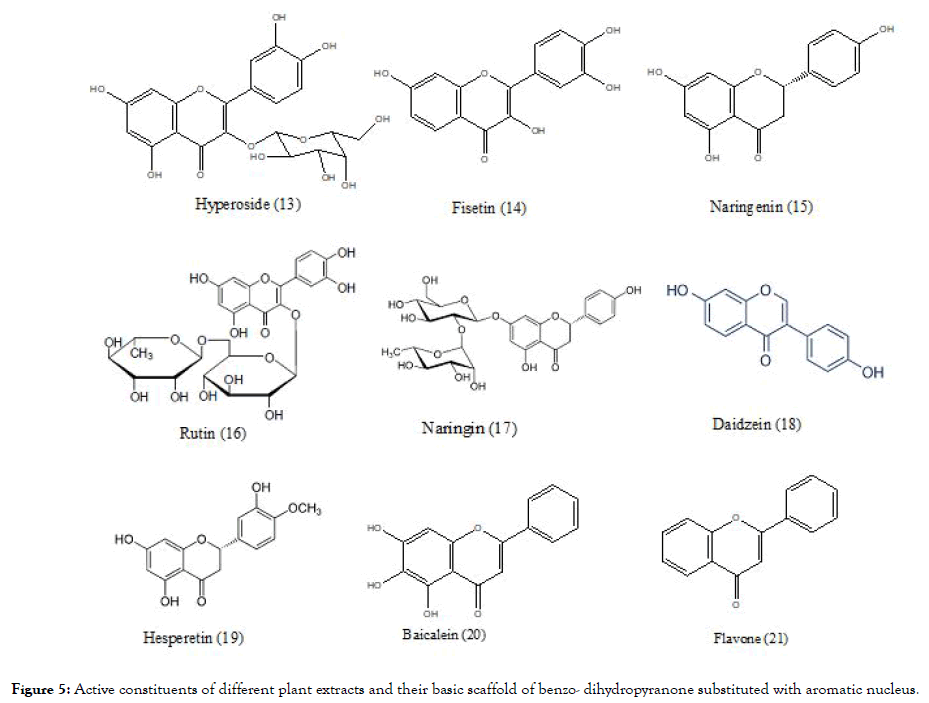
Figure 5. Active constituents of different plant extracts and their basic scaffold of benzo- dihydropyranone substituted with aromatic nucleus.
Zandi et al, evaluated three natural phyto-constituents categorized as flavonoids using in vitro method (Figure 5): fisetin (14), naringenin (15) and rutin (16) against DENV-2; cytotoxic effect (CC50) were studied using Vero cells. The inhibition concentration (IC50) and selectivity index (SI) of fisetin was found to be 55 μg/mL and 4.49 respectively for infected cells and 43.12 μg/mL and 5.72 for uninfected cells, when treated continuously 5hr prior to infection until 4 days’ post infection of virus. Fisetin didn't showed any direct virucidal activity but naringenin exhibited the activity with IC50=52.64 μg/mL, but the SI was low<1 whereas comparing to fisetin and narungenin, rutin didn't showed any significant activity. The overall research work indicates that among three flavonoids, only the fisetin possessed the significant in vitro activity against DENV replication determined experimentally by %RNA reduction level. The mechanism of fisetin is unclear but the investigations suggest that it does not interfere in the binding step of DENV-2 to cells. It might be acting by affecting DENV genome copy number or affecting DENV replication by forming complex with it or acting by interfering replication by affecting RdRp of NS5 protein [80]. Fisetin again complies with the structural scaffold as that of quercetin whereas in naringenin the pyranone ring is saturated which might be responsible for their inactivity. In rutin, the structure deviates from previously discussed hyperoside by having additional glycoside linkage which may contributes to its inactivity.
Zandi [81] also evaluated the flavonoids (Figure 5) quercetin (1), naringin (17), daidzein (18) and hesperetin (19) against DENV-2. They measured DENV replication by Foci Forming Unit Reduction Assay (FFURA) along with quantitative real time polymerase chain amplication (qRT-PCR). Out of four, quercetin exhibited significant inhibitory activity with IC50 35.7 μg/mL. The SI of quercetin was found to be 7.07 for infected cells and 8.74 for uninfected cells, when treated continuously 5hrs before infection until 4 days’ post infection of virus. The exact mechanism by which quercetin exert the activity remains unknown but it is believed to act like other flavonoids by interfering with cellular RNA polymerase and forming complex with RNA [81]. It has again proven by quercetin that the presence of basic scaffold of flavonoid, important for activity. In case of naringin which has structure similarity to naringenin (devoid of saturation in pyranone ring) and rutin (containing glycosidic linkage), indicate the importance of previous structural observations of basic scaffold and also increasing in the glycosidic linkage does not add to the activity as observed in case of rutin. The structure of hesperetin is similar to naringenin and daidzein in which the position of substituted aromatic nucleus to basic scaffold was changed to incorporate the structural diversity might be responsible for their inactivity.
Baicalein (20) obtained from the root of Scutellariabaicalensis- Chinese traditional medicinal herb possessed similar kind of scaffold like flavone (21) (Figure 5). The antiviral activity of baicalein, against various stages of DENV-2 replication in Vero cells are evaluated using FFURA and qRT-PCR. The resultant IC50 value is 6.46 μg/mL for cells prior treated with virus (5hrs) and 5.39 μg/mL post infection upto 4 days and the SI values found to be 17.8 and 21.3 respectively. It also showed IC50=1.55 μg/mL for direct virucidal effect and IC50=7.14 μg/mL for antiadsorption effect. When the scaffold of baicalein compared with that of previous flavone, it indicated that the presence of three hydroxyl group in baicalein, might be useful to its activity [82].
Boesenbergia rotunda (L.) is a common spice of ginger family consisting with flavonoids pinostrobin (22), pinocembrin (23), alpinetin (24), phenylpropanoid cardamonin (25), and its open chain chalcone derivatives pandurantin A (26) and 4-hydroxypandurantin A (27) which are shown in Figure 6. These natural constituents are isolated from above mentioned spice and screened against DENV-2 virus NS2B-NS3 protease by enzyme assay with the use of purified protease [83]. Among all these, pandurantin A and 4-hydroxypandurantin A found to be most active and pinocembrin with least activity. The inhibitory protease activities are shown in Table 1. Interestingly, the compounds pinocembrin (with flavonoids scaffold) and cardamonin (with chalcone scaffold) had low inhibitory effect on NS2B-NS3 protease and showed some synergistic effect when mixed together. Whereas the most active compoundspandurantin A and 4-hydroxypandurantin A which do not have basic scaffold of fused benzopyranone ring, showed competitive inhibitory effect on NS2B-NS3 protease, experimentally determined with inhibitory constant (Ki) of 21 and 25 μM/L, respectively. Interestingly above observations indicate that chalcone, an open chain precursor of flavones as well as flavonoids both showed activity. Panduratin which retains keto benzene ring of the above scaffolds still retains the anti-dengue activity.
| Compound | Percentage inhibition of DENV-2 NS2B-NS3 protease; Concentration used (ppm) | ||
| 120 | 240 | 400 | |
| 23 | 30.1 ± 0.5 | 47.3 ± 0.5 | 56.1 ± 0.4 |
| 25 | 39.4 ± 0.6 | 50.1 ± 0.4 | 71.3 ± 0.3 |
| 23+25 | 52.6 ± 0.4 | 63.5 ± 0.5 | 81.8 ± 0.3 |
| 26 | 87.7 ± 0.6 | 92.2 ± 1.2 | 99.8 ± 1.1 |
| 27 | 87.6 ± 0.4 | 97.3 ± 0.3 | 99.8 ± 0.3 |
Table 1: Percentage inhibition of DENV-2 virus NS2B-NS3 protease cleavage a. The substrate cleaved by NS3 protease used in the experiment corresponded to the peptide Boc-Gly-Arg-Arg-MCA ± corresponds to standard deviation.
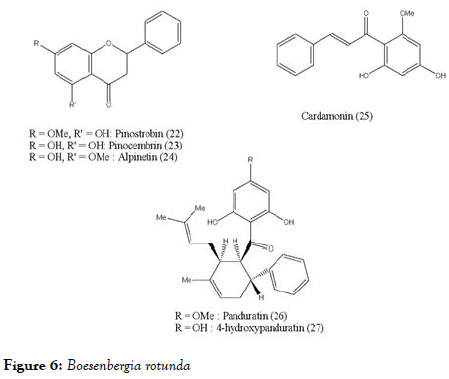
Figure 6. Boesenbergia rotunda
Gynurapseudochina (L) DC, commonly known as Dewa leaves and Curcuma aeruginosa Roxb. Known as temuireng rhizome, were reported to increase the platelet count in patients infected with dengue. The potentiality was determined by using heparin induction and also determined that they did not accelerate the plasma leakage. The main phyto-constituents of dewa leaves are flavonoids, triterpenoids, chlorogenic acid, caffeic acid, vanillic acid, parafumaric acid, p-hydroxy benzoic acid whereas temu rhizome contains flavonoids, alkaloids, polyphenols, curcuminoids and essential oils (tomerone, zingiberene). The dosages concentration of 500 mg/kg body weight (BW) of dewa leave extract; and 250 mg/kg BW and 500 mg/kg BW of temuireng rhizomes extract found to be effective to increase platelet count in patients [84].
Other than flavonoids and chalcones, few alkaloids (Figure 7) also showed the positive activity against DENV. Low and coworkers studied Emetine (28) belonging to the category of ipecacuanha alkaloids identified its activity against dengue at concentration 0.5 μM/L [85]. To determine the mechanism of action of Emetin dihydrochloride, a series of experiments were conducted and concluded that it interfere with viral replication at early stage, either by interfering with the viral RNA synthesis or by affecting the viral protein translation.
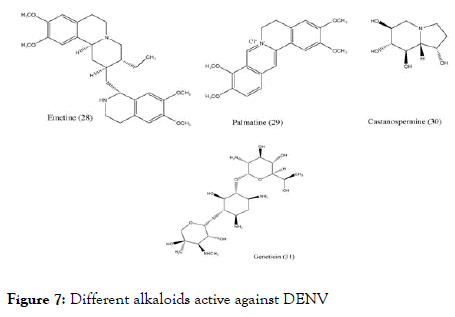
Figure 7. Different alkaloids active against DENV
Another Chinese medicinal plant Coptischinensis Franch containing palmatine (29) showed antiviral activity against DENV-2 in vitro using Vero cells by viral titer reduction assay. Although its EC50 was found to be 26.4 μM/L and the SI to be 39 still its mechanisms of action is not clear [86]. Castanospermumaustrale, main source of Castanospermine (30) tested in vitro and in vivo against all serotypes of dengue. The in vitro studies indicated that it inhibits all serotypes of dengue virus whereas in vivo studies indicated the inhibition of DENV-2. It was found that inhibition occurs at the level of secretion and infectivity of viral particles. Further experiments were carried to understand the effective doses and control on mortality in a mouse model of dengue virus infection, found that the doses of 10, 50, and 250 mg/kg of BW per day were effective in controlling the mortality [87].
Geneticin-aminoglycoside (31) was screened against dengue virus. The in vitro study uses BHK cells for activity, viral plaque reduction assay with western blotting and qRT-PCR for analysis. The outcome of the study concludes that geneticin inhibits DENV-2 proliferation indicated by EC50 of 3 ± 0.1 μg/mL against DENV-2, it decreased the viral yield (EC50 of 2 ± 0.1 μg/mL and EC50 of 20 ± 2 μg/mL), inhibited viral plaque formation in both the number and the size of the plaques (EC50 of 25 μg/mL) and blocked viral RNA and protein synthesis with SI equal to 66 [88]. Interestingly, the studied alkaloid scaffolds showed fused benzo-N-containing heterocyclic ring. They differ in their SI values showing slightly higher selectivity index.
C. laurifolia leaf extract has been screened and recognized as a hit against the DENV- NS5 RdRp. The bioguided fractionation of the leaf extract (Figure 8) lead to five flavonoids, namely quercetin (1), 6-methylapigenin-7-methylether (32), avicularin (33), quercitrin (34) and hyperoside (13), along with betulinic acid (35). The activity of fractionated compounds was compared with commercial flavonoids: isoquercitrin (36), spiraeoside (37), quercetin-3,4'-di-O- glucoside (38) and rutin (16). Compounds quercitrin, betulinic acid, spiraeoside and rutin displayed IC50 ranging from 1.7 to 2.1 μM, and were the most active against the DENV-NS5 RdRp. Among tested compounds betulinic acid were originated as stronger inhibitor of NS5 RdRp with IC50 as 1.7 μM (Table 2) and it was first reported triterpene against dengue. It will be fascinating in the upcoming days to evaluate the activity of other triterpene derivatives on the DENV-NS5 [89]. Interestingly, hyperoside showing significant EC50 value in the previously reported experiment, showed higher IC50 value in inhibiting DENV-NS5 RdRp.
| Compound | IC50 | Structural features |
|---|---|---|
| Quercetin | 3.6 | Fused Benzo-dihydropyranone ring |
| Hyperoside | >20 | Fused Benzo-dihydropyranone ring with one glycosidic unit |
| Rutin | 2.1 | Fused Benzo-dihydropyranone ring with two glycosidic units |
| Avicularin | 1.7 | Fused Benzo-dihydropyranone ring with one glycosidic unit |
| Quercitrin | 2.1 | Fused Benzo-dihydropyranone ring with one glycosidic unit |
| Betulinic acid | 1.7 | Triterpene |
| Spiraeoside | 1.9 | Fused Benzo-dihydropyranone ring, glycosidic unit on aromatic nucleus |
Table 2: Inhibition of DENV-NS5 RdRp (IC50 in μM) by compounds from C. laurifoliaand other related commercial flavonoid monomers.
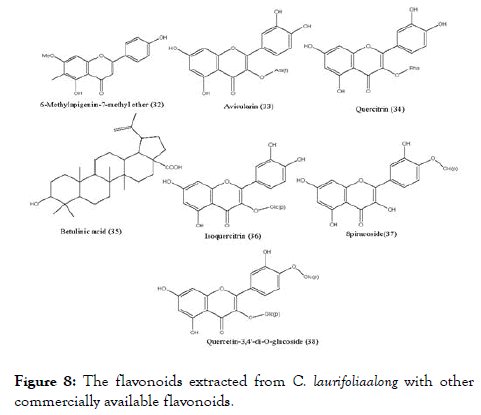
Figure 8. The flavonoids extracted from C. laurifoliaalong with other commercially available flavonoids.
The various parts of Cryptocaryachartacea plant were used to prepare different ethyl acetate extracts using 650 new Caledonian plants to screen their biological activity against NS5 polymerase. Enzyme assays were performed and several active extracts were found but among them the most active one is from the bark of Cryptocaryachartacea Kostern belonging to the family Lauraceae. This extract was used for isolation and further separated non-alkylated flavonoid pinocembrin (23) as well as series of chartaceones A-F (39-44) (Figure 9) consisting structural feature based on new mono and dialkylated ones. Out of screened compounds, chartaceones C-F (41-44) had shown IC50 ranging from 1.8 to 4.2 μM/L against DENV-2 NS5 polymerase. On the contrary, compound pinocembrin was completely inactive. These results signify the importance of alkylated chains in the structures of chartaceones C-F, which may play an imperative role in inhibition of polymerase [90].
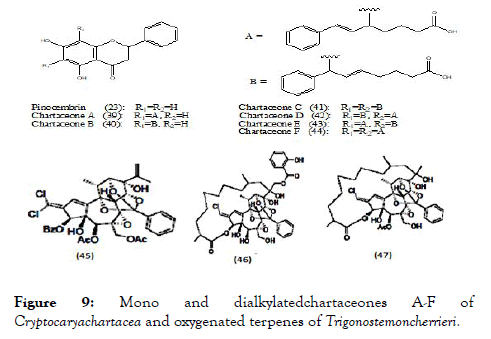
Figure 9. Mono and dialkylatedchartaceones A-F of Cryptocaryachartacea and oxygenated terpenes of Trigonostemoncherrieri.
Trigonostemoncherrieri, the bark and the wood of this rare plant when explored for their chemical composition, resulted in the isolation and characterization of several oxygenated terpenes, among them compounds (45-47) (Figure 9) were evaluated against DENV RdRp and showed IC50 of 12.7 ± 0.2, 3.1 ± 0.2 and 16.0 ± 1.3 μM/L respectively [91].
However, while exploring the natural phyto-constituents in search of novel anti-dengue agent, it was observed that scaffolds require specific structural features such as fused benzodihydropyranone ring or benzo-N-heterocyclic rings substituted with aromatic nucleus. Many times additional glycosidic substituent or triterpene nucleus indicated along with main scaffold. Overall these features prevail in the scaffolds targeted especially towards NS2B-NS3 and NS5 RNA polymerase.
Synthetic scaffold-targeting NS2B-NS3 and NS5
Based on the natural scaffolds, many synthetic molecules have been designed and screened against DENV or evaluated for specific target. Various scaffolds targeted towards NS2B-NS3 and NS5 are discussed here. Lai et al., synthesized and studied a series of functionalized 1,2-benzisothiazol-3(2H)-one-1,3,4- oxadiazole (48) (Figure 10) hybrid derivatives and were subsequently screened against DENV and West Nile virus proteases. Out of twenty-four compounds, ten showed promising IC50 values against DENV2. The IC50 value of compound (48) against DENV2 was found to be 3.75 ± 0.06 μM. The kinetic studies for the same revealed a competitive mode of inhibition [92]. The scaffold (48) consists of fused benzo- thiazole with heterocyclic aromatic ring substitutions.
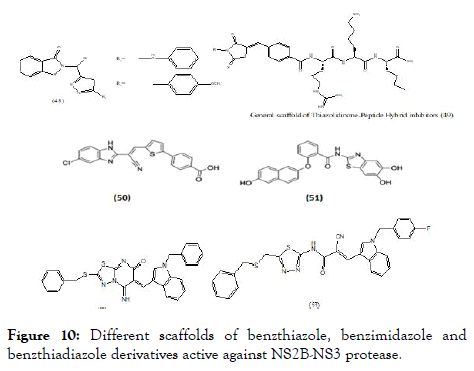
Figure 10. Different scaffolds of benzthiazole, benzimidazole and benzthiadiazole derivatives active against NS2B-NS3 protease.
Klein et al. reported molecular hybridized peptide derivatives against more specific dengue NS2B-NS3 protease. The structural features more focuses on the heterocyclic moiety with incorporation of N-substituted 5-arylidenethiazolidinone as N-terminal capping groups of the peptide moiety. The general scaffold contains non fused thiazole with amide (peptide) type substitution (49), (Figure 10). The in vitro study of these molecules showed binding constant (Ki) values between 1.5 and 1.8 μM and indicated competitive inhibition mechanism. The studies revealed that the hybridization of peptide with thiazolidinedione along with substitution of different hydrophobic groups showed most promising activity [93]. Raut screened an in-house library and revealed molecule with benzimidazole scaffold (50), as a potent inhibitors of DENV-2 protease with an IC50 value of 5.95 μM and at 30 μM showed significant reduction of viral titers in all four serotypes [94]. Wu et al. reported compound (51), as inhibitor of DENV-2 and 3 protease with IC50=4.2 μM and IC50 =0.99 μM respectively [95]. A cell-based protease assay revealed an IC50 of 3.2 μM. This compound also showed promising antiviral activity in Vero cells (EC50=0.8 μM) but had cytotoxic effects at concentrations above 10 μM. Liu et al. explained about heterocyclic molecules based on thiadiazoloacrylamide (Figure 10) against NS2B-NS3 protease and from which thiadiazolopyrimidinone (52) reported as a new chemical structure against protease complex. Based on the above observation thiadiazo nucleus was further modified to a series of analogs. From the series, fourteen analogs were identified as potent protease inhibitors. Considering the structural features of these series, revealed the importance of nitrile group linkage for inhibitory activity. The best of these (53) demonstrated an IC50 at 2.24 μM based on in vitro DENV2 NS2B-NS3 pro assays [96].
Doman et al. virtually screened 3 lakh compounds using Autodock 3 in search of hit compound against the NS2B-NS3 protease, from the screening data selected thirty six compounds and evaluated them using in vitro assay for the protease inhibition. Out of thirty six compounds, three strong inhibitors (54-56), (Figure 11) were showed competitive inhibition with Ki values of 4.0 ± 0.4 μM (IC50=15.6 ± 0.9 μM), 4.9 ± 0.3 μM (IC50=12.5±0.5 μM), and 3.4 ± 0.1 μM (IC50=3.9±0.6 μM), respectively [97].
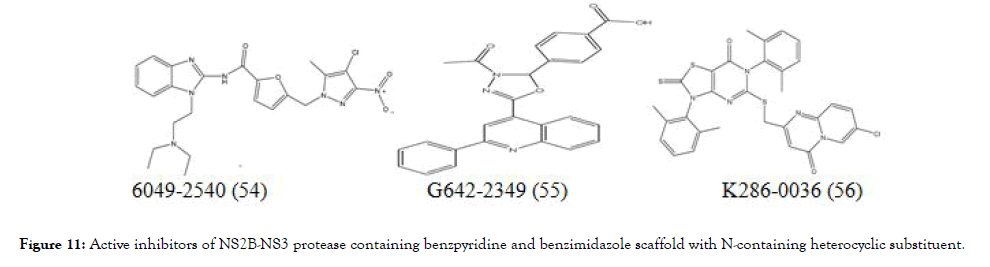
Figure 11. Active inhibitors of NS2B-NS3 protease containing benzpyridine and benzimidazole scaffold with N-containing heterocyclic substituent.
Interestingly the most active inhibitors identified to have Ncontaining heterocyclic scaffold such as benzpyridine and benzimidazole. Although aryl substituted thiazolopyrimidine showed activity but it was comparatively at lesser extent than the above.
Some research groups also separately studied the inhibitory effect on NS3 helicase activity (Figure 12). In this regard Ivermectin (57), reported as antiparasitic drug, also identified activity against NS3-WNV helicase using in silico docking. In biochemical assay it displayed inhibition of only helicase unwinding activity of NS3 by uncompetitive mode of action with an EC50 of 0.7 μM for DENV [98]. The same compound did not show any activity towards DENV helicase ATPase. Basavannacharya et al. performed High throughput screening using a molecular beacon helicase assay and found suramin against DENV helicase, as a noncompet itive inhibitor with a Ki of 0.75 μM [99]. In addition to above mentioned inhibitors acting against DENV NS3-helicase unwinding activity, the reports also suggest activity of few inhibitors towards ATPase activity. Li et al. [100] synthesized a series of inhibitors based on dimeric benzothiazole scaffold, which were further studied by Ndjomou et al. [101] revealing in vitro inhibitors of DENV helicase ATPase activity. The compound (58) observed with highest DENVhelicase as well as helicase unwinding activity with an IC50 of 0.5 μM and 1.5 μM respectively and in DENV replicon assay had an EC50 value of 7.1 μM [102]. Similarly observed in this series, primuline analogs against both RNA unwinding and ATPase activities of NS3. Best of Pyrrolone derivative (59) showed precise activity against ATPase as well as inhibition of DENV replication with EC50=36 μM but has poor selectivity (CC50/EC50=4.5) [102].
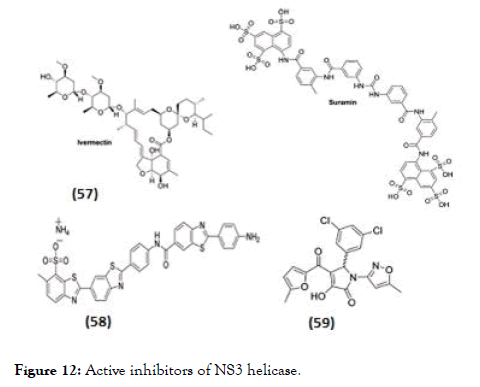
Figure 12. Active inhibitors of NS3 helicase.
NS5 is another important non-structural protein studied extensively with various synthetic scaffolds targeting either its methyltransferase domain (Figure 13) or towards RNA (RdRp) polymerase domain (Figure 14). Designing NS5 methyltransferase and guanylyltransferase inhibitors are gained importance currently. Two methyltransferase competitive inhibitors which presently available are SAH, a product inhibitor of the methylation reaction, the natural product sinefungin, and S-adenosyl-L methionine (SAM)-a nucleoside analogue of the methyl donor [103,104]. These compounds are nonselective inhibitors with activity against viral and eukaryotic methyltransferases. Whereas in case of sinefungin has inhibitory effect on both guanine N-7 (IC50 values=0.030 μM) as well as the ribose 2'-O MTase (IC50 values=0.041 μM) with greater significance in IC50 values compared to SAH (IC50 values of 1.77 and 0.49 μM) [104]. The higher inhibitory activity related towards its binding affinity, since sinefungin displayed a 6-fold higher Ki value in comparison to SAM (136 vs. 826 nM) [105]. A series of molecules having 1,2,3-triazole scaffold derived from 3'-azidothymidine (AZT) proven for HIV-1 inhibition, the same series were modified by adding a bulky silyl protecting group (TBS) at the 5' position [106] and evaluated against DENV MTase inhibition. From the series, molecule (60) shown promising activity in reducing viral titre by 98% at 10 μM with EC50=7.4 μM but the selectivity was poor [106]. Docking studies of these molecules, suggested that the binding of the 5'-TBS group to the hydrophobic cavity near the SAM binding site [106]. A HTS discovered 2-thioxothiazolidin-4-ones (rhodanines) as a potent inhibitor of DENV NS5 GTP-binding and guanylyltransferase activity [107]. In a DENV replicon assay, (61) identified as a promising inhibitor of NS5 guanylation, with an IC50 and an EC50 of 30.8 μM.
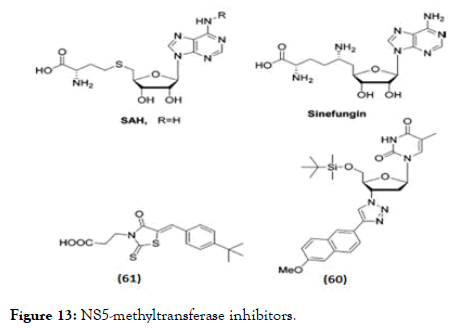
Figure 13. NS5-methyltransferase inhibitors.
NS5 RNA polymerase inhibitors are classified as nucleoside inhibitors (NI) and non- nucleoside inhibitors (NNI) depending upon their acting site (62-70), (Figure 14). NI directly bind to the polymerase site and act as RNA chain terminators whereas the NNI act at allosteric sites of NS5, thus inhibiting its enzymatic activity. For the biological activity, the NIs of DENV polymerases need to be converted into their active triphosphate form with the help of cellular kinases. Therefore, the synthesis of the corresponding nucleoside triphosphate (NTP) is essential to study their activity in biochemical assays [108].
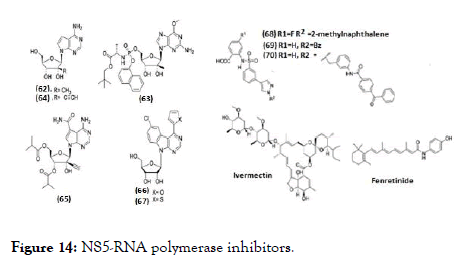
Figure 14. NS5-RNA polymerase inhibitors.
The development of RNA polymerase inhibitors progressed with screening of known HCV NIs against DENV. 7-Deaza-2'-Cmethyl- adenosine (62), evaluated against DENV-2 using AG129 mice model and found with EC50=15 μM [109, 110]. Similarly, another HCV NI (63), prodrug of 6-O-methyl-2'- C-methylguanosine has EC50=14.2 nM against DENV-2 [111]. Compound (64) is a 7-deaza-2'-C-ethynyl analogue of adenosine, inhibited DENV serotypes 1-4 in cell- culture, potency concentration ranges from micro to sub-micromolar level. The biologically active triphosphate form competes with the natural substrate resulting in subsequent chain termination in the RNA polymerase assay [112]. 65 is an ester prodrug of 64, chemical modifications were associated with either lower activity or higher cytotoxicity but the compound was not further pursued because of observed toxicity in rats [113-115]. Nucleoside analogues containing a class of benzo-fused 7-deazapurine scaffold revealed inhibitory activity against DENV in cell culture within micromolar to sub-micromolar concentration [29,116] (Figure 15). Optimization to the bioisosteric 2-thienyl derivative (67) from potent but cytotoxic 2-furyl analogue (66), almost retained the potency and yielded a higher selectivity index (EC50=0.335 μM in Huh-7, CC50=19.92 μM in HepG- 2) [117]. Compounds (68) and (69) are the first reported allosteric inhibitor of RNA polymerase. Compound (68) is more potent with IC50=0.26 μM while compound (69) with IC50=7.2 μM [117]. In another series, benzophenone derivative (70) was evaluated and is anticipated to block template entry and act on NS5 RdRp by allosteric binding mechanism [117,118]. It was the only derivative which showed effect in a cell-based plaque assay, where it reduced DENV titer by 3.4 and 4.5-fold at 6 and 17 μM, respectively. Ivermectin and fenretinide belongs to NNI class and identified as inhibitors of the NS5 by an alpha-screen assay. It acts by interacting with importin (IMP)-α/β1, thus blocking NS5 nuclear localization [119-121]. Ivermectin along with its activity against NS3, it also prevents NS5 binding to IMP- α/β1 with an IC50 of 17 μM [121] and inhibited DENV 1-4 with EC50 of 1.2-1.6 μM in the CFI assay [120]. Whereas fenretinide or 4- HPR (N-(4-hydroxyphenyl)-retinamide) is a synthetic retinoid, reported to affect steady-state DENV genome replication [122] and was reported to modulate the unfolded protein response (UPR) by activating the protein kinase R-like endoplasmic reticulum (PERK) arm [121]. Further, 4-HPR prevented the association of NS5 to IMP- α/β1 and IMP αIBB in an alphascreen assay with IC50 values of 1-1.5 μM and reduced viral replication in cell culture for DENV serotypes 1-4 and ADE infection by comparable EC50 values (0.8-2.6 μM) [121,122].
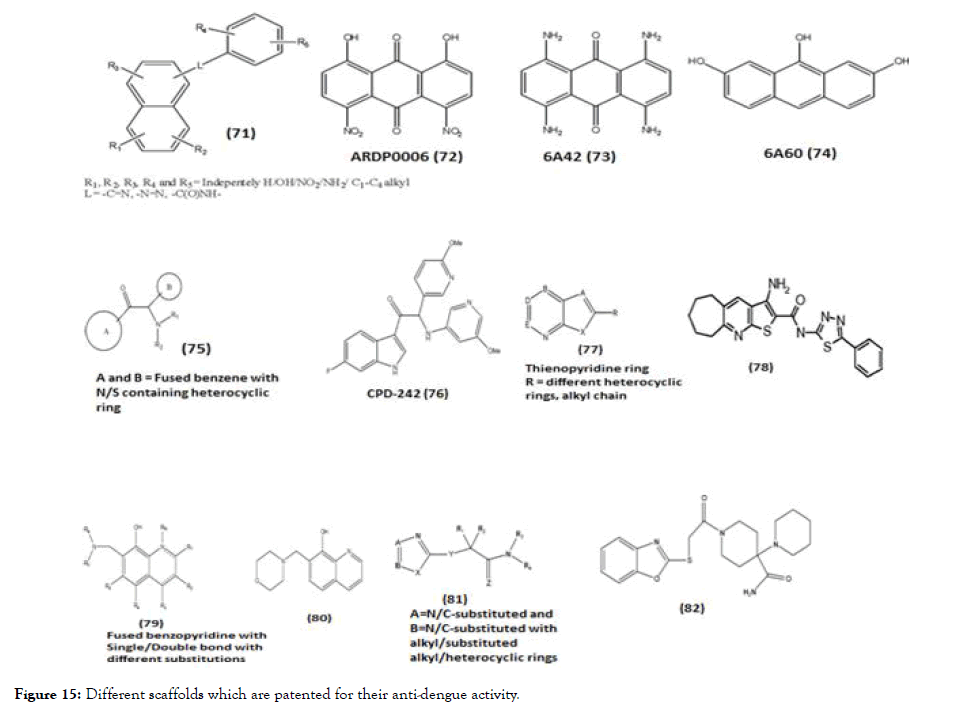
Figure 15. Different scaffolds which are patented for their anti-dengue activity.
Patented scaffolds-targeting NS2B-NS3 and NS5
In this review, we briefly describe about some of the general scaffolds which are patented for DENV where the reported activity is targeted towards NS2B-NS3 and NS5. Compound (71) is a scaffold with polycyclic nucleus like anthracene substituted with various substituents [123]. The enzyme kinetic studies of synthesized molecules belonging to this scaffold 6A42 (73) and 6A60 (74) showed the NS2B-NS3 protease activity like the parent molecule ARDP006 (72). Interestingly the kinetic inhibition data describes ARDP0006 as competitive inhibitor while the analogues were best described as mixed, noncompetitive inhibitors. This patent also described the NS2BNS3 activity of DENV2 for the certain lead molecules such as Ivermecetin, selamectin, haematoxylinpentaacetate, tyrothricin and alexidine, they were otherwise known for their different activities such as anti-parasite, topical broad spectrum parasiticide, transacetylase inhibitor, antibiotic and anticancer agent respectively. Except tyrothricin, all lead molecules showed mixed inhibitor activity while tyrothricin showed competitive inhibition [123]. Another patent scaffold (75) and its derivatives include A and B ring connected with amino-ethanone linkage, evaluated against DENV serotype 1-4 and the EC50 and CC50 of all compounds were determined. The A ring mostly fused benzofive/ six membered heterocyclic ring and B ring consists of substituted phenyl/benzene/heterocyclic ring/fused heterocyclic ring. The in vitro studies were conducted on the above compounds against NS3 and NS5 using RT-PCR. Among the series of synthesized molecule, CPD-242 (76) were further selected for in vivo study of DENV-2 strain and it was found that CPD-242 led to highly significant delay in virus-induced morbidity [124]. Patent containing thienopyridine scaffold (77) and their derivatives evaluated against DENV2 along with all other serotypes. Compound 1 (78) of the series was identified as one of the most potent molecule while almost all analogues of thienopyridine scaffold were active against dengue with EC50 values at or below 25 μM [125]. Adding to this, patent claimed for various scaffolds, out of which benzopyridine scaffold (79) and its various substituted derivatives were evaluated for DENV2 replicon inhibition and cytotoxicity. In the series, compound 33 (80) showed the highest therapeutic index (TI) compared to other derivatives [126]. Patent consists of benzoxazole (81) derivatives and analogs with various substituents like alkyl/aryl/heterocyclic rings. Twenty-two compounds were identified as hits showing EC50 value below 25 μM, out of these hits, compound (82) was identified as most potent with activity against all four serotypes of dengue [127]. Adding to these, patent based on 2,4- diaminoquinazoline nucleus (83) with different alkyl/aryl/heterocyclic substitution tested for DENV-2 and their IC50 values protease enzyme were determined using replication method, where compound Yhhu-1036 (84) showed the lowest IC50 value 0.009 μM [128].
The current review article covers the major perspective of research work related to DENV with respect to molecules from naturals, synthetic source and some of molecules from patents. Since many years scientists throughout the world are collaborating with their expertise and striving hard to find out the hit molecules to treat DENV, a life threatening disease. All the scaffolds which are screened till date signify that majority of molecules predominately shows nitrogenous heterocyclic rings or fused benzene ring with different hetero atoms especially nitrogen and or sulfur. Also substitutions of functional groups like halogens, hydroxyl or amino further enhances their electronic environment favorable for binding interactions with various targets. This review article thus will surely be a great stepping stone to highlight significance of various new scaffolds that can specifically inhibit the various targets of the dengue.
The current review article covers the major perspective of research work related to DENV with respect to molecules from naturals, synthetic source and some of molecules from patents. Since many years scientists throughout the world are collaborating with their expertise and striving hard to find out the hit molecules to treat DENV, a life threatening disease. All the scaffolds which are screened till date signify that majority of molecules predominately shows nitrogenous heterocyclic rings or fused benzene ring with different hetero atoms especially nitrogen and or sulfur. Also substitutions of functional groups like halogens, hydroxyl or amino further enhances their electronic environment favorable for binding interactions with various targets. This review article thus will surely be a great stepping stone to highlight significance of various new scaffolds that can specifically inhibit the various targets of the dengue.
Citation: Ganji LV, Kanyalkar MA (2019) Non-structural Proteases as a Target of Dengue Virus. J Antivir Antiretrovir. 11:188. DOI:10.35248/1948-5964.19.11.188
Received: 05-Oct-2019 Accepted: 15-Oct-2019 Published: 21-Oct-2019
Copyright: © 2019 Kanyalkar MA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.