Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2021)Volume 14, Issue 12
Several protein domains and receptors recognize minimotifs on the short carboxyl termini of proteins and control protein clustering, trafficking, and posttranslational modifications. These functional C-terminal minimotifs are found on approximately 17% of proteins in the human proteome, with 83% of unknown functional significance. We tested a bioinformatic/proteomic approach to systematically identify new C-terminal minimotif functions. We selected 30 putative consensus C-terminal minimotifs based on their fold-enrichment and sequence complexity. These 30 consensus minimotifs had instances on 16% of the C-termini in the human proteome. Binding partners for a representative instance for each consensus C-terminal minimotif were identified by affinity purification liquid chromatography-tandem mass spectrometry. We validated the approach with a PDZ binding C-terminal minimotif instance, SDV> and identified 436 potential new interactors. For 7 of the 30 new C-terminal minimotifs, 32 previously known interactors were rediscovered. Overall, the experiments support 2,048 new binding partners to the 30 C-terminal minimotifs. Many interactors of the LxxxI> and QxxL> minimotif sequence patterns have role in RNA splicing and cell cycle, respectively. The key consensus residues for the new minimotifs were at positions of pathogenic mutations for 6 diseases giving insight into disease mechanism. In conclusion, the 30 new C-terminal consensus minimotifs cover 16% of the human C-terminome, help identify potential disease mechanisms, and this approach can be scaled to determine functions of protein C-termini in the human C-terminome.
Minimotif; Short linear motifs; Mass spectrometry; Proteomics; C-terminome; C-terminus; Consensus sequence; Peptide; TAP-tag; Spliceosome; Splicing, Proteome; Protein-protein interaction; Mental retardation; Deafness; Hyper-IgM syndrome; Neurodevelopmental disorder with microcephaly; Impaired language; Gait abnormalities; Osteogenesis imperfect; Antithrombin III deficiency
Sets of similar conserved short strings of amino acids (minimotif instances) can be used to derive consensus minimotifs for proteinprotein interactions, protein trafficking, and post-translational modifications (PTM) [1-3]. Identification of these consensus sequences has contributed vastly to our understanding of protein function and regulation. While minimotifs are located anywhere within a protein, a small subset is only functional when located at the Carboxyl-terminus [1,4]. For instance, the peroxisomal targeting signal 1 is sufficient for protein trafficking to peroxisomes when present at the C-terminus [5-9]. The KDEL receptor binds to [H/K] DEL sequence on the end of endoplasmic reticulum (ER) resident proteins. This motif is a signal to retrieve Endoplasmic Reticulum (ER) proteins that escape to distal compartments of the secretory pathway [10-13]. Deleting or blocking the KDEL> sequence by extending the C-terminus results in the secretion of mutated ER proteins establishing necessity. Appending the C-terminus of proteins otherwise destined to be in other subcellular compartments, with the KDEL> sequence can retain the proteins in the ER demonstrating sufficiency [10,14]. The majority of the PDZ domains bind to Class I (x-[S/T]-x-Φ>), II (x-Φ-x-Φ>), and III (x-[DE]-x-Φ>) minimotifs present exclusively at the C-terminus; however, there are exceptions. These interactions are essential for cellular signaling [15]. Nonsense or missense mutations in the C-terminal minimotifs can disrupt protein function and lead to diseases [4,16]. Several examples include a mutation in the VxPx> minimotif of Rhodopsin protein causes autosomal dominant retinitis pigmentosa, deletion of the C-terminus in human HERG channels causes long QT syndrome type 2 (LQT2). Although only a few are known, these diseases indicate the significance of C-terminal minimotifs to human health [17-19].
Several cell processes including alternative splicing, multiple translational stop-sites, and proteolysis increase the number and diversity of unique C-terminal ends [4]. It is estimated that a cell might have a 100,000-500,000 unique C-termini at any given time. The ProTEUS database contains novel C-terminal sequences [20-22]. Mass-spectrometry-based approaches and in silico prediction of new C-termini arising from proteolysis and alternative splicing are some of the approaches to identify new C-termini in the TopFIND database [23-25].
Although there are significant efforts to identify novel C-termini, far less work has focused on the unique functions of the C-termini. The molecular functions of only 3,500 C-terminal minimotifs are supported by some experimentation [1]. This fraction is only 17% of the human proteome. We previously consolidated functional information into the human C-terminome database [1]. In our original C-terminome paper several computational strategies predicted new minimotif instances:
1. Verified consensus sequences;
2. C-terminal minimotifs identified in rodent proteomes and conserved in human paralogs; and
3. Predictions of over-represented C-terminal sequences and patterns in the human proteome. Even with these approaches 82% of C-termini in human proteome do not have a known molecular function.
We are interested in exploring strategies to address this problem. However, we must consider some limitations. The small size, limited binding surface, and generally weaker binding affinity of minimotifs make it challenging to determine the molecular functions of C-terminal minimotifs [4]. Additionally, residue degeneracies in some positions of minimotif consensus sequences are often revealed through alanine scanning or other mutagenesis experiments. These experiments are labor-intensive and time-consuming, thus not scalable to the proteome level. High-throughput methods such as peptide microarrays, protein microarrays, peptide phage display, yeast 2-hybrid, and yeast cell surface display are a mainstay for the study of modular minimotifdomain interactions [26]. Proteomic approaches are routinely used to find new PTM instances [27]. Like the proteomic approaches used do analyze cell extracts, we sought an approach that could assess minimotif interaction in a native mammalian cell context.
In this paper, we tested an approach of computationally screening for candidate minimotifs followed by affinity mass spectrometry. As part of the computational approach, we previously generated and searched for all possible sequence patterns on the C-termini of human proteins. These putative minimotifs were searched for over-represented sequences in the human proteome to infer a conserved function. Candidate C-terminal minimotifs were further evaluated by Tandem Affinity Purification (TAP)-tag strategy to isolate C-terminal minimotif binding partners and complexed proteins in Liquid Chromatography tandem massspectrometry (LC-MS/MS)-based approach. We reasoned that by testing the patterns of degenerate positions of higher fold enrichment in the human proteome, we can deduce the molecular functions of putative C-terminal minimotifs for a larger set of proteins. Our results suggest that this approach can be scaled to evaluate the functional C-terminome.
Generation and prioritization of predicted C-terminal minimotifs
A detailed method for generating predicted C-terminal sequence patterns and sequences was as described previously [1]. Briefly, we downloaded the RefSeq protein database of human proteins. Anchored sequence patterns and instances of length 3-10 amino acids were generated with at most five degenerate positions for the last ten amino acids of all human proteins. Fold enrichment for each sequence pattern, and the instance was calculated as previously described [1]. Thirty sequence patterns were selected based on their fold enrichment and sequence complexity (Figures S1 and S2). For each selected C-terminal pattern, a list of proteins matching the pattern was retrieved from the RefSeq database. A single representative protein was randomly chosen, and the last ten amino acids were tested by affinity LC-MS/MS.
TAP-tagged C-termini expression vectors
To build the TAP tag fused C-terminal motif expression vectors, we first designed and built the vector backbone expressing the TAP tag. The TAP-tag is a codon-optimized two IgG-binding domains from Protein A, a TEV protease cleavage site, nine contiguous Myc tags, and a stop codon. The pUC.TAP cDNA was PCR amplified, digested with Nhe1/EcoR1 enzymes, and the cDNA was ligated into pcDNA3.1(-) Myc/HisA using Quick Ligase (New England Biolabs, Ipswitch, MA, Cat. #: M2200S). The ligation mixture was transformed into DH5α competent cells. The plasmid was purified using a Qiagen miniprep kit (Qiagen, Hilden, Germany) and the insert was confirmed by DNA sequencing.
To clone the C-terminal motifs in frame with TAP tag, we built the oligo duplexes as follows. Single-stranded oligonucleotides with flanking Sac2 and EcoR1 restriction sites were synthesized at Sigma Aldrich and IDTdna. Each sequence and its reverse complement encoded the selected last ten amino acids of one of 30 proteins containing a potential C-terminal minimotif (Table S1 and S2). Each oligonucleotide was phosphorylated by incubating with 10 mM ATP, 10X kinase buffer, and T4 polynucleotide kinase (NEB cat # M0201S) for 4 hr at 37°C. The reaction mixture was heat-inactivated at 65°C for 20 minutes. Reverse complementary oligonucleotides (3.2 μM) were annealed by incubating at 45°C for 10 min, and cooled to 25°C in thermocycler TC5000, producing oligonucleotide duplexes for subcloning.
The oligonucleotide duplexes were cloned in frame with the TAP tag into a pcDNA3.1(-1) Myc/HisA vector at the Sac2 and EcoR1 restriction sites. This process was used to build the 30 expression constructs, each containing a different C-terminal minimotif. For each experimental C-terminal minimotif, a control with conserved consensus residues mutated to alanine was constructed. Individual clones were sequence-verified (Beckman Coulter, Indianapolis, IN; Genewiz, South Plainfield, NJ; and GenScript, Piscataway, NJ). The TAP-tag vector was used as an empty vector control expressing only the epitope tags.
Additional control vectors were designed and built by overlap PCR (Figure S3). The modifications included increasing the length and change in the sequence of a linker region followed by TEV protease cleavage site. Another modification was changing the TEV cleavage site to sites for Thrombin and PreScission protease. The PDZ domain was amplified through RT-PCR from cDNA obtained from pEAK Rapid cells and ligated in frame with another TAP-tag vector containing HA and FLAG epitope tags.
Co-immuno precipitation and TCA precipitation
pEAK Rapid cells (passages 1-10) were seeded at 2.2 × 106 cells/100 mm dish one day prior to transfection. Each pcDNA. TAP construct (5 μg) were transfected into four 100 mm dishes using LipofectamineLTX with Plus Reagent (ThermoFisher, Waltha, MA, Cat. #: 15338100). After culturing for 48 hours, cells were rinsed with warm filtered PBS and harvested in 2 ml/ dish ice-cold lysis buffer (50 mM HEPES, pH 7.6, 150 mM NaCl, 10% glycerol, 1% NP-40, 0.5% Triton X-100 with 500 μL of freshly prepared 30 mg/ml PMSF and protease inhibitors). Cells were chilled on ice for 5 min and centrifuged at 16,873 x g for 15 min at 4°C to remove cellular debris. Total protein content of supernatants was measured through a bicinchoninic acid protein assay (Pierce, Appleton, WI, Cat. #: 23227).
For co-immunoprecipitation, 5 mg of total cell lysate was incubated with a monoclonal Myc antibody (DSHB hybridoma, University of Iowa, Iowa City, Iowa; 9E10) in lysis buffer for 2 hr at 4°C with shaking. The antibody-cell lysate mixture was incubated with 50 μL of Protein G-magnetic bead conjugates (BioRad, Hercules, CA, Cat. #: 161-4023) for 30 min at 4°C. The beads were washed twice in lysis buffer followed by an additional two washes in the lysis buffer lacking detergent. The bound antibody complexes were eluted twice with 200 μL of glycine buffer (100 mM glycine, pH 2.0) incubating for 15 min at 20°C with gentle mixing. The eluate was neutralized by mixing with an equal volume of 500 mM Tris, pH 8.0. The final pooled eluate was divided into three aliquots; 10% of the eluate was analyzed on protein gel to confirm the Co-IP. Another 10% was saved, and 80% of the eluate was subjected to label-free LC-MS/MS after precipitation with TCA.
For TCA precipitation, the eluate was incubated with 60% volume of 33% TCA on ice for 30 min. The suspension was centrifuged at 16,873 x g for 15 min at 4°C. The pellet was washed two times with 200 μL iof ce-cold 100% acetone centrifuging at 4°C for 5 min. Pellets were air dried and shipped to the W.M. Keck MS and Proteomics Facility (Yale University, New Haven, CT) for LC-MS/MS. Protein expression was confirmed by Western blot analysis. After glycine elution, precipitates were resuspended two times with 50 μL 1x Laemmli buffer and heated at 95°C for 2 min. The Laemmli eluates were precipitated with 100% ice-cold acetone. The elution was incubated in a 4x volume of 100% icecold acetone at 4°C for at least 2 hr. Samples were centrifuged at 16,873 x g for 15 min at 4°C. The samples were washed again, and the pellet was air-dried, and LC-MS/MS was conducted at the W.M. Keck MS and Proteomics Facility. All experiments were performed in triplicate. For co-immunoprecipitation experiments using IgG beads, approximately 2 mg of the total cell lysate was incubated with 50 μg of IgG beads, complexes were eluted with 100 mM glycine, pH 2.0 elution buffer and processed as described above.
TAP-tag immunoprecipitation
500 μL of total cell lysate was incubated with 50 μL of IgG beads (GE Healthcare Life Sciences, Marlbourough, MA, Cat. #: 17- 0969-01) for 2 hr. Beads were washed three times with lysis buffer and exchanged into TEV protease cleavage buffer. Beads were incubated with TEV protease at 4°C overnight. Samples were centrifuged at 16,873 x g for 15 min at 4°C, and the supernatant was incubated with 500 μL of spent Myc hybridoma (9E10) culture media. The precipitated Myc-C-terminal peptide complexes were isolated by incubation with 50 μL of Protein G beads (GoldBio, St. Louis, MO, Cat. #: P-430-1), washed to remove any unbound proteins, and protein complexes were removed in 60 μL of the 1x Laemmli buffer incubating at 95°C for 5 min. Protein expression was confirmed by Western blotting. The following antibodies were used: Prot-A (GenScript, Cat. #: A01778), and HA (Santa Cruz Biotechnology, Santa Cruz, CA, F-7, Cat. #: sc7392).
LC-MS/MS analysis of SxxxK>, VxxL>, and VxxxS> putative C-terminal minimotifs
We first tested three of the thirty putative C-terminal minimotifs to establish an overall approach. Immunoprecipitated pellets were resuspended in 100 μL 50 mM ammonium bicarbonate and protein was measured with an EZQ Protein assay (Molecular Probes, Eugene, OR, Cat. #: R33200). Samples were reduced in 5 mM dithiothreitol) and alkylated with 15 mM iodoacetamide. These samples were digested with 1 μg trypsin/Lys C (Promega, Cat. #: V5071) overnight at 30°C. Resulting peptide mixtures were separated using an UltiMate 3000 RSLCnano system (Thermo Scientific, San Jose, CA) with a self-packed UChrom C18 column (100 μm × 35 cm). Peptides were eluted with a 90 min gradient of solvent B from 2-27% (solvent A: 0.1% formic acid; solvent B: acetonitrile, 0.1% formic Acid) at 50°C using a digital Pico View nanospray nozzle (New Objectives, Woburn, MA) that was modified with a custom-built column heater and an ABIRD background suppressor (ESI Source Solutions, Woburn, MA). The self-packed column tapered tip was briefly pulled with a laser micropipette puller P-2000 (Sutter Instrument Co, Novato, CA) to an approximate id of 10 μm. The column was then packed with 1-2 cm of 5 μm Magic C18 followed by 35 cm of 1.8 μm UChrom C18 (120A) at 9000 psi using a nano-LC column packing kit (nano LCMS, Gold River, CA).
The mass spectral analysis was performed using an Orbitrap Fusion mass spectrometer (Thermo Scientific, San Jose, CA). Proteomic analysis was performed using a “Universal” datadependent method as per the Thermo Application note. The MS1 precursor selection range was from 400-1500 m/z at a resolution of 120,000, and an intensity threshold of 4.0 × 105. Quadrupole isolation at 0.7 Th for MS2 analysis using CID fragmentation in the linear ion trap with a collision energy of 35%. The automatic gain control was set to 1.0 × 102 with a maximum injection time of 250 ms. The instrument was set in a top speed data-dependent mode with a most intense precursor priority. Dynamic exclusion was set to exclusion duration of 60 s with a 10-ppm tolerance.
MS/MS chromatograms were extracted, and the charge state was deconvoluted with Proteome Discoverer version 2.1. All MS/ MS samples were analyzed with SEQUEST (ThermoFisher, San Jose, CA, USA; version 2.0.0.802). SEQUEST was configured to search a custom C-terminal peptide fasta, assuming the digestion enzyme Trypsin and max number of missed cleavages set to 2. SEQUEST searched were set with a fragment ion mass tolerance of 0.60 Da and a parent ion tolerance of 10.0 PPM. Variable modifications included carbamidomethyl of cysteine, oxidation of methionine, deamidation, and acetylation of the N-terminus. MS/MS-based peptide and protein identification was validated with Scaffold (version Scaffold_4.8.2, Proteome Software Inc., Portland, OR). Peptide identifications were accepted if they could be established at greater than 80.0% probability by the Peptide Prophet algorithm with a Scaffold delta-mass correction. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least two peptides from the same protein. Protein probabilities were assigned by the Protein Prophet algorithm [28]. Proteins that contained ambiguous peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
LC-MS/MS analysis of a positive control and 27 putative C-terminal minimotifs
Protein pellets were dissolved and denatured in 10 μl of 8 M urea with 0.4 M ammonium bicarbonate. The proteins were reduced by the addition of 1 μL 45 mM dithiothreitol (Pierce Thermo Scientific Cat. #: 20290) and incubated at 37ºC for 20 min. Denatured proteins were alkylated by after the addition of 1 μL 100 mM iodoacetamide (Sigma-Aldrich, St. Louis, MO, Cat. #: I1149) and incubation in the dark at 20ºC for 20 min. The urea concentration was adjusted to 2 M by the addition of 27 μL of water. Proteins in samples were digested with 0.5 μg of trypsin (Promega, Madison, WI, Cat. #: V5113) for 16 hr at 37ºC. Samples were desalted using C18 Ultra microspin columns (The Nest Group, Ipswitch, MA Cat. #: SUM SS18V) following the manufacturer’s directions eluting peptides with 0.1% TFA, 80% acetonitrile. Eluates were dried on a Speedvac and dissolved in MS loading buffer (2% acetonitrile, 0.2% trifluoroacetic acid). Protein concentration and purity (A260/A280) was assessed with determined with a Nanodrop Spectrophotometer (Thermo Scientific, Nanodrop 2000 UV-Vis). Each sample was diluted with MS loading buffer to a concentration of 0.04 μg/μL, with 0.2 μg (5 μL) injected into the LC-MS/MS instrument. For some samples 0.4 μg was loaded: SxV> (IgG, Myc-ProtG), AxS>, ExP>, ExxA>, ExxxS>, GxxK>, LxxK>, LxxxF>, LxxxI>, PxxK>, PxxS>, QxxL>, and RxxR> (0.4 μg injected).
LC-MS/MS analysis was performed at the Keck Proteomics Facility on a Thermo Scientific Orbitrap Fusion equipped with a Waters nanoAcquity UPLC system utilizing a binary solvent system (Buffer A: 100% water, 0.1% formic acid; Buffer B: 100% acetonitrile, 0.1% formic acid). Trapping was performed at 5 μL/ min, 97% Buffer A for 3 min using a Waters Symmetry® C18 180 μm × 20 mm trap column. Peptides were separated using an ACQUITY UPLC PST (BEH) C18 nanoACQUITY Column 1.7 μm, 75 μm × 250 mm (37°C) and eluted at 300 nL/min with the following gradient: 3% buffer B at initial conditions; 6% B at 5 min; 35% B at 170 min; 50% B at 175 min; 97% B at 180 min; 97% B at 185 min; return to initial buffer conditions at 186 min.
MS was acquired in the Orbitrap in profile mode over the 350- 1,550 m/z range using quadrupole isolation, 1 microscan, 120,000 resolution, AGC target of 4E5, and a maximum injection time of 60 ms. MS/MS chromatograms were collected in top speed mode with a 3 s cycle time on species with an intensity threshold of 5E4, charge states 2-8, peptide monoisotopic precursor selection preferred. Dynamic exclusion was set to 45 s. MS/MS data were acquired in the Orbitrap in centroid mode using quadrupole isolation, HCD activation with a collision energy of 28%, 1 microscan, 60,000 resolution, AGC target of 1E5, the maximum injection time of 100 ms.
LC-MS/MS data were analyzed using Proteome Discoverer (version 1.3) software and searched with the Mascot algorithm (version 2.6.0) (Matrix Science, Chicago, IL). The data were queried against the SwissProt database with the taxonomy restricted to Homo sapiens and the database was customized with the TAP-tag minimotif bait sequences. Search parameters included trypsin digestion with up to 2 missed cleavages; peptide mass tolerance of 10 ppm; MS/MS fragment tolerance of +0.02 Da; and variable modifications of methionine oxidation and carbamidomethylated cysteine. Normal and decoy database searches were searched, with the confidence level set to 95% (p<0.05).
Experimental design and statistical rationale
Post LC-MS/MS, resulting files were analyzed using Scaffold 4.0 free version at 1% peptide and 1% protein false rate discovery threshold with minimum of two peptides. The files were analyzed for total protein probability. The annotated human protein information from the UniProt website (http://www.uniprot. org/) was queried for the UniProt ID, Domain [CC] (DOMAIN_ CC), Domain [FT] (DOMAIN) [29]. A program in Perl was written to parse the results downloaded from the above UniProt queries. The parsing results were used for internal queries to retrieve structural information for a list of proteins with UniProt IDs obtained from a common C-terminal minimotif. Finally, the retrieved structural information was summarized for each list of proteins containing the common minimotif. Count numbers on each structural domain name for every C-terminal minimotif were calculated for structural domain profiling.
We identified disease-associated mutations in new C-terminal minimotifs. All data were collected, cleaned, and analyzed with custom Python scripts, utilizing the pandas, numpy, lxml, and re libraries. The releases of the ClinVar XML file and the FASTA files from RefSeq were the current versions as of April 30, 2020. The ClinVar database was analyzed to select variants that mutated a key amino acid in one of the new consensus C-terminal minimotifs [30,31]. Each missense and nonsense singlenucleotide variant was matched to a RefSeq protein, including multiple isoforms, to identify the reference and alternate amino acids in the C-termini [32]. Allele frequencies and counts for each variant were matched from gnomAD Exomes v2.1.1 [33]. After cleaning the data, the reference and alternate amino acids in the C-termini were matched with the 30-consensus sequence regular expressions to identify variants that change a key amino acid in the C-terminal consensus minimotif.
We developed a strategy to discover new C-terminal minimotifs with molecular functions (Figure 1). The first step in our workflow was to identify the putative C-terminal minimotifs for experimentation (Figure 1A). In the example presented here, for an artificial proteome, a consensus minimotif pattern is defined as the conserved sequence elements in a group of multiple single instances found in the proteome. An example of a consensus sequence is PxP> for which there are four instances present in this artificial proteome. Putative minimotifs were selected based upon their enrichment in the human proteome when compared to random synthetic proteomes (Table S1). For each consensus minimotif we randomly selected a protein that contains the C-terminal pattern (Figure 1B).
For the selected minimotif instances we generated constructs to express fusion proteins with the last ten amino acids containing the C-terminal minimotif (Table S2). Negative controls had either empty vector or minimotifs with Ala substitutions for each of the key consensus amino acids. This approach was chosen to maximize sensitivity to identify C-terminal minimotifs, rather than proteins that bind to some region of the C-terminus. Minimotifs are short and generally have weaker affinities than other protein-protein interactions. Each plasmid was transfected in mammalian cells and epitope tagged C-terminal motif was enriched from lysates by affinity purification. The epitope tagged C-terminal minimotif was eluted along with its binding partners (endogenous proteins) and the samples were analyzed by massspectrometer to identify potential binding partners (Figure 1C). Data was analyzed with MASCOT and/or Sequest software. The resulting peptides were queried against the human database to identify the potential proteins and to an internal database of the bait proteins. Identified interactors were analyzed with Scaffold software. Background proteins or non-specific interactors in the vehicle or Ala mutant controls were removed. The identified proteins were assessed with BioGRID, UniProt, and SMART databases to identify their physiological relevance to the C-terminal minimotifs.
Profile of the selected putative C-terminal minimotifs
We postulated that identifying the function of a sequence pattern instead of an instance sequence will be more broadly applicable to multiple proteins (Figure 1). Consequently, we analyzed the human C-terminome database. The database contains putative C-terminal sequence patterns present in the human proteome (1). We selected 30 such patterns from this set for experimental testing based on their fold enrichment score, length, sequence complexity, and number of instances in the human proteome (Figure S1 and Table S2). The experimentally verified C-terminal minimotifs in the human C-terminome database have a mean fold enrichment score of >1.0 and a mean length of seven amino acids. Based on this, the mean and the highest fold enrichment scores of selected predicted patterns were 1.4 and 9.0 respectively. The minimotif lengths varied between 3-5 amino acids. The 30 selected consensus motifs had 5,578 instances at the C-termini of proteins in the human C-terminome, covering about 16% of the human proteome.
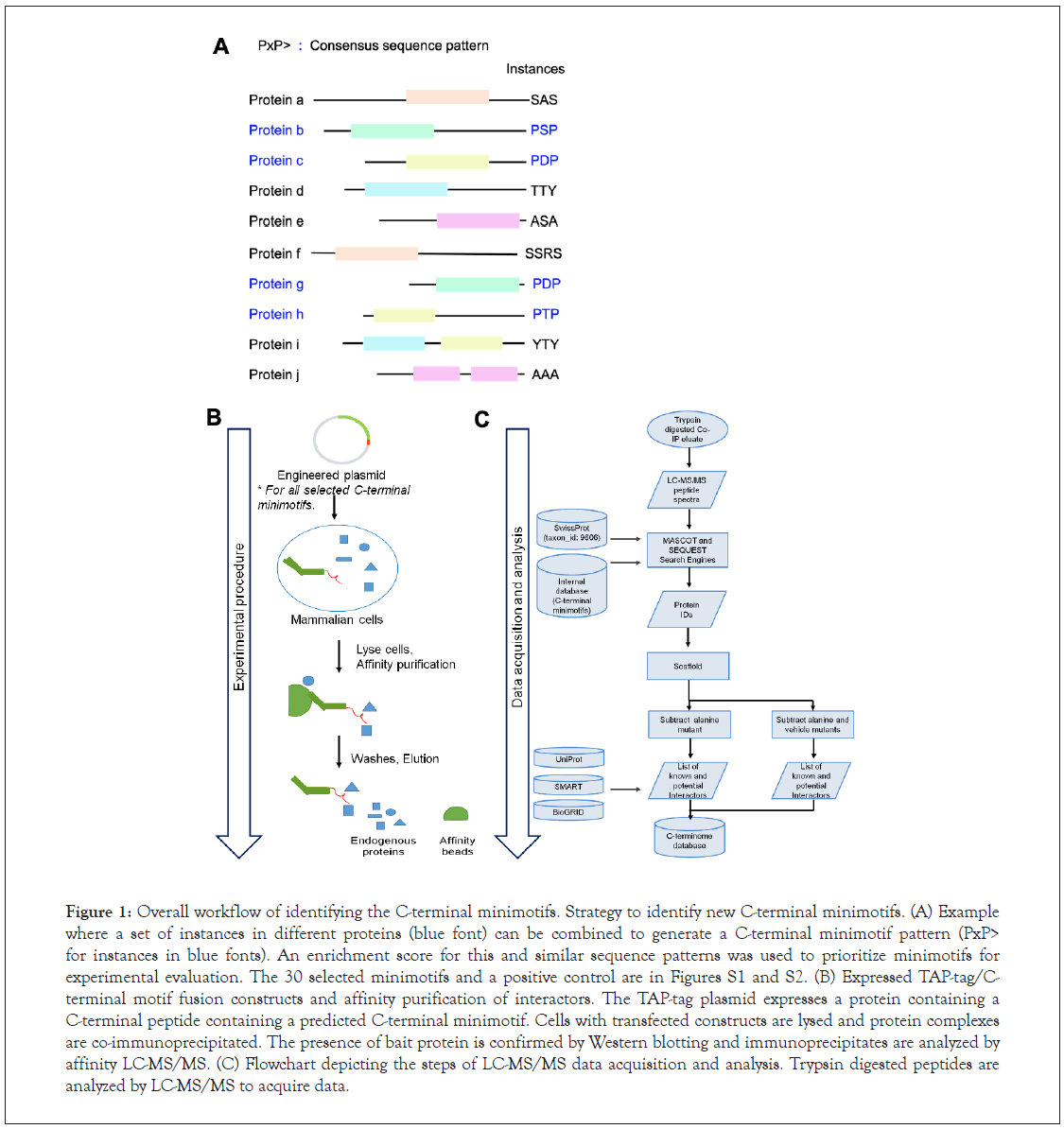
Figure 1: Overall workflow of identifying the C-terminal minimotifs. Strategy to identify new C-terminal minimotifs. (A) Example where a set of instances in different proteins (blue font) can be combined to generate a C-terminal minimotif pattern (PxP> for instances in blue fonts). An enrichment score for this and similar sequence patterns was used to prioritize minimotifs for experimental evaluation. The 30 selected minimotifs and a positive control are in Figures S1 and S2. (B) Expressed TAP-tag/Cterminal motif fusion constructs and affinity purification of interactors. The TAP-tag plasmid expresses a protein containing a C-terminal peptide containing a predicted C-terminal minimotif. Cells with transfected constructs are lysed and protein complexes are co-immunoprecipitated. The presence of bait protein is confirmed by Western blotting and immunoprecipitates are analyzed by affinity LC-MS/MS. (C) Flowchart depicting the steps of LC-MS/MS data acquisition and analysis. Trypsin digested peptides are analyzed by LC-MS/MS to acquire data.
The consensus residues and degeneracies of the 30 minimotif patterns in the human C-terminome were visualized with Logoplots (Figure S2) [34,35]. We aligned the last 10 amino acids from the C-terminus of proteins for each sequence pattern. Only amino acids for the original pattern were conserved, suggesting that the selected C-termini did not contain other minimotifs. The 30 minimotif sequence instances were selected from different subcellular localizations, although most were from the cytoplasm and/or nucleus (60%).
Validating the minimotif pull down strategy with a known C-terminal minimotif
TAP is a two-step purification approach reducing the non-specific interactors in the eluted fractions. It is based on the principle that the two epitope tags in tandem are separated by a protease cleavage site such that the incubation with the protease and pulldown with the second epitope tag with successive purification steps reducing background. Our first TAP-tagged protein could not be cleaved at the protease cleavage site despite altering salt, detergents, and denaturants in our lysis buffer (Figure S3A). These experiments suggested that steric hindrance blocked access to the protease cleavage site. To alleviate this problem, we built variations of our initial TAP-tag plasmid by altering the length and the sequence of the linker C-terminus to the protease cleavage site as well as modifying the cleavage site for different protease specificity (Figure S3, B-E). Changing the linker length and the sequence had no impact on cleavage. However, exchanging the protease cleavage site for Thrombin and Precision Protease released the second epitope tag. Despite extensive efforts, we could only recover small amounts of cleaved protein, which were suitable for the downstream analysis. Therefore, we proceeded with a onestep purification protocol.
We tested the well-documented C-terminal minimotif and PDZ domain interactions as a positive control [36]. PDZ are wellcharacterized domains of approximately 100 amino acids in length, and function as scaffolds for protein-protein interactions with ion channels and other signal proteins [36]. While there are many subclasses of PDZ domain binding motifs, for the purpose of a positive control, we chose a class I minimotif pattern (x[S/ T[x[L/V]>) [37]. The representative sequence for the pattern was SDV> from the N-methyl-D-aspartate (NMDA) receptor, known to bind the PDZ domain of the post-synaptic density (PSD-95) protein [38]. We co-immunoprecipitated cell lysates co-expressing a TAP-tag fusion protein containing SDV> minimotif and a HA-tagged PDZ domain. As expected, the PDZ domain bound its cognate minimotif, minimal binding to the negative control minimotif with alanine substitutions for key consensus residues was observed (Figure 2). This experiment confirmed that a TAPtagged C-terminal minimotif can immunoprecipitate its binding domain.
Figure 2: Testing the pull-down approach with a known C-terminal SxV> minimotif and an epitope tagged interactor with a PDZ domain. The SDV> instance of SxV> C-terminal motif is present in GRIN2A protein and the PDZ domain is present in the PSD-95 protein. Both SDV> and PDZ domain were cloned and co-expressed in pEAK Rapid cells. Co-immunoprecipitation experiments were performed on pEAK Rapid cell lysates using IgG beads co-expressing FLAG.HA. conjugated PDZ domain and TAP-tag fusion protein containing SDV> minimotif (labeled as SxV>). Shown here is the Western blot confirming the interaction between the SxV> minimotif and a PDZ domain. TAP-tag only (i.e. no C-terminal sequence) and TAP-Cterm alanine substitution mutant (AxA>) were the two negative controls.
To determine whether novel interaction partners could be identified, eluates of SDV> minimotif were examined by LC-MS/ MS under varied experimental conditions, MS instrumentation, and software (Table S3). MASCOT searches produced a larger number of predicted proteins overall when compared to that of SEQUEST searches (Table S3 and Figure S4). Both lists were analyzed for potential interactors and for PDZ domain containing proteins at different statistical thresholds (Tables 1, S4 and S5). After removing the false positives, we looked for PDZ domain containing proteins that are known to interact with the SDV> minimotif at varying thresholds (Table 1). No PDZ domain-containing proteins were discovered at a conventional statistical threshold (triplicates observations) at 1% protein and 1% peptide false discovery rate with a minimum number of 2 peptides) (Table S6). However, Afadin was in all samples, and has a PDZ domain that binds to class I PDZ binding minimotif [39] (Table S3). Afadin was detected in most conditions.
| Samples/Beads2 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antibody | Myc | None | Myc | Myc | Myc(Dil) | Myc(Dil) | Myc #1 | Myc #2 | Myc #3 | |||
| Thresholds | Search engine | Protein names | Instrument | Fusion | Fusion | Fusion | Qep | Fusion | Qep | Fusion | Fusion | Fusion |
| Beads | ProtG | IGG | Agrose ProtG | Magnetic ProtG | ||||||||
| Protein IDs | Probability | |||||||||||
| 1% protein and peptide FDR, minimum # of | MASCOT | Afadin | AF AD | 0% | 0% | 37% | 100% | 0% | 0% | 0% | 0% | 0% |
| 1% protein and peptide FDR, minimum # of | MASCOT | Afadin | AF AD | 0% | 0% | 36% | 100% | 0% | 0% | 0% | 0% | 0% |
| Afadin | AF AD | 0% | 0% | 0% | 0% | 0% | 0% | 10% | 95% | 96% | ||
| 5% Protein and peptide FDR, minimum # of peptides: 2 | MASCOT | Membrane- associated granulate kinsase, WW and PDZ domain-containing protein 1 |
MAGI 1 | 0% | 0% | 36% | 100% | 0% | 0% | 0% | 0% | 0% |
| Afadin | AF AD | 0% | 0% | 36% | 100% | 0% | 0% | 10% | 95% | 96% | ||
| Lim domain only Protein 7 | LMO 7 | 0% | 0% | 7% | 0% | 0% | 0% | 0% | 0% | 0% | ||
| 5% Protein and peptide FDR, minimum # of peptides: 1 | MASCOT | Membrane- associated granulate kinsase, WW and PDZ domain-containing protein 1 | MAGI 1 | 0% | 0% | 36% | 100% | 0% | 0% | 0% | 0% | 0% |
| PDZ domain-containing protein 11 | PDZ 11 | 0% | 0% | 0% | 0% | 0% | 96% | 0% | 0% | 0% | ||
| Golgi reassembly-stacking protein 2 | GORS 2 | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 84% | 0% | ||
| 90% protein and 90% thresholds, minimum# of peptides: 1 | MASCOT | Afadin | AF AD | 0% | 0% | 36% | 100% | 0% | 0% | 0% | 0% | 0% |
| 1% protein and peptide FDR, minimum # of | SEQUEST | Afadin | AF AD | NI | NI | 55% | 0% | 0% | 0% | NP | NP | NP |
| 5% protein and peptide FDR, minimum # of | SEQUEST | Afadin | AF AD | NI | NI | 55% | 0% | 0% | 0% | NP | NP | NP |
| 1% protein and peptide FDR, minimum # of | SEQUEST | Afadin | AF AD | NI | NI | 55% | 0% | 0% | 0% | NP | NP | NP |
| 5% protein and peptide FDR, minimum # of | SEQUEST | Afadin | AF AD | NI | NI | 55% | 0% | 0% | 0% | NP | NP | NP |
| 90% protein and 90% thresholds, minimum# of peptides: 1 | SEQUEST | Lim domain only Protein 7 | LMO7 | 98% | 0% | 0% | 0% | 0% | 0% | NP | NP | NP |
| Afadin | AF AD | NI | NI | 55% | 0% | 0% | 0% | NP | NP | NP |
Note: NP: Not Processed; NI: None Identified; Dil: diluted sample. List of PDZ domain containing proteins identified after removing the proteins identified in the respective alanine mutant negative control.
Table 1: Domain analysis of the proteins identified through LC-MS/MS screen on the SxV>, a PDZ binding minimotif.
Afadin is an adherence junction scaffolding protein that binds with the all classes of PDZ domain binding minimotifs [39]. In particular, Afadin PDZ domain binds with the Breakpoint cluster region protein (Bcr) containing TEV> minimotif [40]. TEV>, like the SDV> minimotif, is also a class I PDZ binding minimotif. As we decrease the stringency, other PDZ domain-containing proteins were detected such as MAGI1, LMO7, PDZ11, and GORS2 (Table 1). The human papillomavirus E6 oncoprotein binds to the PDZ domain of MAGI1-through its x(T/S)xV>, a class I PDZ binding minimotif [34]. This minimotif interaction is essential for the proteosome-mediated degradation of MAGI1 [41]. The minimotif interacting with LMO7, PDZ11, and GORS2 is not known. We also identified some novel binding partners of C-terminal peptide containing the SxV> minimotif that do not have a PDZ domain and need further experimentation for validation (Tables S4 and S5). These results establish the experimental conditions and analysis parameters for analyzing other minimotifs and validate this approach to identify additional minimotif functions (Figure 3).
Figure 3: Number of immunoprecipitated proteins identified in the affinity LC-MS/MS screen for each C-terminal minimotif. Putative C-terminal minimotifs expressed in pEAK Rapid cell lysates were immunoprecipitated to identify their endogenous binding partners by LC-MS/MS. The number of proteins identified in the LC-MS/MS screen for each minimotif after removing background proteins that bind to controls.
We applied the most stringent criteria to reduce false positives and the false discovery rate, thereby producing higher confidence results. Additionally, we analyzed our samples on the Fusion LCMS/ MS instrument because it was more sensitive in capturing the low abundant proteins when compared to the QEp instrument. We also noticed that co-immunoprecipitation with Myc antibody resulted in identification of more immunoprecipitated proteins when compared to the proteins immunoprecipitated using IgG beads. Therefore, for testing putative C-terminal minimotifs, we selected the above-mentioned conditions to decrease the background.
Interactors of putative C-terminal minimotifs
We systematically analyzed the data obtained from LC-MS/MS chromatograms. We first confirmed the presence of the bait protein in the searches. To do so, we checked the percentage coverage of the full-length TAP-tag protein encoding putative C-terminal minimotif and the 15 amino acids from the C-terminus of the TAP-tag proteins. The presence of bait proteins containing the C-terminal minimotif confirmed that the interactors are likely to be co-immunoprecipitated through the C-terminal minimotifs.
Minimotif-domain interactions are important for cell signaling. Approximately 75% of the ~10,000 structural protein domain families have no known binding minimotif sequence [42].
Therefore, we further analyzed our results targeted toward discovering novel C-terminal minimotif and protein domain interactions. Since ExxA>, QxxL>, and LxxxI> minimotifs each co-immunoprecipitated more than 100 proteins, the minimotifs were more likely to have multiple interactors with a common domain (Table S7). We examined the immunoprecipitated proteins for these three minimotifs for over-represented domains when compared to the alanine mutants and empty negative controls (Tables S8). Consistent with the positive SDV> control, several potentially unique C-terminal minimotifs: protein domains interactions were observed.
We determined if the pull-downs for each minimotif had previously known interactions with the protein with the protein C-terminal minimotif instance. Known interactors of the 30 proteins were downloaded from the BioGRID database. For the 30 C-terminal minimotifs studied (Tables S1 and S2); we identified 32 known interactors of the endogenous proteins for seven putative C-terminal minimotifs (Table 2 and Tables S9-S34). At least five of these interactors have been previously confirmed by the low-throughput experiments such as yeast two-hybrid or co-fractionation. The rest of the interactions were previously identified through affinity LC-MS/MS, which will require additional validation experiments. Some interactors were also present in the empty vector negative controls (Table S35-S40).
| Minimotif sequence | Known interactors | Potential interactors | Potential Interactors (Based on GO Biological Function) | ||
|---|---|---|---|---|---|
| WT- Mut | WT- (Mut+vehicle) | WT- Mut | WT- (Mut+vehicle) | WT- (Mut+vehicle) | |
| QxxL | 0 | 0 | 229 | 205 | 20 |
| PxxS | 0 | 0 | 60 | 11 | 1 |
| AxS | 0 | 0 | 66 | 53 | 0 |
| LxxxF | 17 | 14 | 115 | 96 | 2 |
| ExP | 0 | 0 | 10 | 6 | 0 |
| GxxK | 0 | 0 | 5 | 3 | 0 |
| PxxK | 3 | 3 | 235 | 212 | NA1 |
| RxxR | 0 | 0 | 12 | 10 | 0 |
| SxxxK | 2 | 0 | 79 | 14 | 2 |
| VxxL | 0 | 0 | 107 | 7 | 2 |
| VxxxS | 1 | 0 | 191 | 6 | 0 |
| ExxA | 3 | 0 | 538 | 47 | 3 |
| LxxxI | 3 | 1 | 110 | 32 | 10 |
| ExxxS | 0 | 0 | 26 | 5 | 1 |
| LxxK | 3 | 1 | 15 | 9 | 0 |
| RxxxV | 0 | 0 | 71 | 55 | NA |
| AxxK | 0 | 0 | 56 | 5 | 0 |
| SxA | 0 | 0 | 31 | 15 | 0 |
| AxP | 0 | 0 | 18 | 2 | 0 |
| RxxxL | 0 | 0 | 18 | 4 | 0 |
| LxT | 0 | 0 | 10 | 4 | 0 |
| GxxxL | 0 | 0 | 9 | 6 | 1 |
| PxxF | 0 | 0 | 9 | 2 | 0 |
| RxxP | 0 | 0 | 6 | 3 | 0 |
| RxxxP | 0 | 0 | 6 | 3 | 0 |
| KxS | 0 | 0 | 5 | 2 | 0 |
| LxxxA | 0 | 0 | 3 | 2 | 0 |
| FxS | 0 | 0 | 2 | 2 | 0 |
| SxK | 0 | 0 | 6 | 5 | 0 |
| Total | 32 | 19 | 2048 | 826 | 42 |
Note: NA1: Needs analysis.
Table 2: Summary Table of co-immunoprecipitation/ LC-MS/MS analysis of predicted C-terminal consensus minimotifs.
Although these published interactors support our approach; it was not previously known that the C-terminal minimotif was a determinant for the interaction. Thus, our results infer a mechanism for the protein interactions mediated through the carboxyl end.
After removing the background by subtracting the common proteins between the alanine mutant, and empty vector negative controls (Tables S35 and S36), we identified 2,048 potential minimotif binding partners summarized in (Table 2). 10% of the putative minimotifs had hundreds of interactors, suggesting that we had indeed identified a novel consensus sequence (Figure 4). Less than 10 interactors were identified for the PxxS>, LxxK>, RxxP>, AxxK>, PxxF>, GxxK>, ExxxS>, GxxxL>, RxxxL>, LxxxA>, SxK>, LxT> and ExP> patterns, suggesting that these minimotif consensi may not be functionally relevant in multiple pathways. No interactor was noted for RxxR> pattern. For FxS>, KxS>, RxxxP>, AxP>, and SxA> patterns, several interactors belonged to the protein families that appear commonly in the negative controls such as IgGs and keratins and are likely to be false interactors. To further evaluate the relevance of 2,048 immunoprecipitated proteins, we matched their GO term with that of the bait minimotif protein. 49 of these interactors had a GO term matching 11 putative C-terminal minimotif sequences (Table S7). Of the 30 tested patterns, LxxxI> and QxxL> minimotif patterns were of the highest interest because the majority of interactors in each set had specific GO pathways.
Figure 4: Percentage coverage of the human C-terminome with a known molecular function. Pie-chart showing the percentage of functional human C-terminome identified by different computational or experimental approaches. Published C-terminal minimotif sequences based on the literature review (17%), inferences based on the experiments done in rodents (<1%), and predictions based on experimentally derived consensi (1%) were categorized previously in the human C-terminome by our group (1). Of the untested over-represented C-terminal patterns (82%), we are predicting functions for an additional <1% proteome and identify candidate functions for 16% of the proteome based on the results LC-MS/MS approach.
The LxxxI> minimotif occurs in 174 proteins in the human proteome with a fold enrichment of 1.2 (Table S1). The LKKSI> instance from the dual specificity protein kinase CLK1 isoforms 1 and 2 was used in the pull-down. CLK1 is a nuclear protein that auto-phosphorylates itself and phosphorylates serine, threonine and tyrosine residues on its protein substrates. Substrates of CLK1 are SRSF1, SRSF3, and PTPN1. SRSF1 and SRSF3, in particular, are SR proteins that have a role in RNA splicing [43,44]. Analysis of the affinity LC-MS/MS results of the CLK1 minimotif identified Bclf1, a previously known interactor of CLK1 [45] and 32 additional interactors. Ten of these proteins either directly or indirectly affect RNA splicing (Table S41). These are CD11A, GCFC2, T2FB2, GPAM1, PCF11, IWS1, RNPS1, TR150, CL043, and CA052 (Table S41). RNA splicing by the spliceosome produces multiple transcripts expressing different protein isoforms. There are over 200 proteins in spliceosomes. However, only a small subset of these proteins has been functionally characterized. Thus, any new information can help to better understand its mechanism [46].
The QxxL> minimotif is found in 181 proteins in the human proteome and has a fold enrichment of ~1.1 (Table S1). We used the QNHL> instance in Nek11 isoform 1 (1). Nek11 is a G2/M checkpoint associated kinase. During DNA damage, Nek11 phosphorylates CDC25A, a CDK activator, inducing its degradation by the proteosome and consequentially cell cycle arrest [47]. Given the role of Nek11 in DNA damage response, cell cycle arrest and DNA replication, many of the proteins in the NEK11 immunoprecipitates had matching GO terms. Twenty proteins (CDC16, CAF1B, KIF4A, GNAI3, CNDH2, DCAF1, DIP2B, DNLI1, MSH3, DPOD1, PSF2, PSF3, 3MG, RPAC1, KIF2C, MDIL1, STT3B, GRDN, HS2ST and SPC24) (Table S42) had matching cell cycle and DNA replication functions.
Potential new minimotif mutations that are pathogenic.
Previous studies have identified several mutations in C-terminal diseases that are pathogenic in humans [1]. We searched the ClinVar database to determine if any of the 20 new minimotifs consensus sequences had mutations in the ClinVar database with corresponding allele frequencies in GnomAD suggestive of pathogenesis. There were 5,591 new minimotifs that were eliminated by a truncating nonsense mutation on the C-terminus of proteins, 2,730 of which were pathogenic for a disease.
While these could potentially be due to loss of a minimotif, we next focused in on variants that eliminated a key consensus residue in a minimotif. There were 146 variants in the new C-terminal consensus residues, however most were benign, likely benign, or variants of uncertain significance; only 6 were pathogenic variants (Table S43). The potentially pathogenic consensus minimotifs were mutated in GXXXL, PXXK, RXXP, SXK, SXXXK, and VXXL minimotifs in the proteins coded by the SERPINC1, CD40LG, NARS1, CRYM, TUSC3, and COL1A1 genes, respectively (Tables 3 and S43). For those variants appearing in GnomAD, the allele frequencies were relatively rare (1.6 × 10-5 to 5.5 × 10-6), frequencies like other pathogenic mutations. The diseases associated with these SNPs were Antithrombin III deficiency, Hyper-IgM syndrome, Neurodevelopmental disorder with microcephaly, impaired language, and gait abnormalities, Deafness, Mental retardation, and Osteogenesis imperfecta. The identification of disease-associated variants with a C-terminal consensus sequences, identifies a potential molecular basis for the pathogenic disease mechanism.
| Clinical significancere | Relationship preferrece | Relationship | Traitpreferredname | Preferred symbol | Duplicate variant | Combined Pr | Ref C Termini | Alt C Termini | Grch37chr | Af | Ac | An | Minimotif |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pathogenic | Crystallinmu | CRYM | Deafness autosomal dominant 40 | DFNA40 | K,314,T | NP_001879 | LIYDSWSSGK | LIYDSWSSGK | 16 | NULL | NULL | NULL | SXK |
| Pathogenic | CD40ligand | CD40LG | Hyper -leg M ayndrome type 1 | NULL | G,257,DC | NP_000065 | GFTSFGLLKL | GFTSFGLLKL | X | 5.52E-06 | 1 | 181044 | GXXXL |
| Pathogenic | asparag inyl -tRNAsynth | NAR51 | Neurodevelopmental disorder with Microcephaly | NEDMILG | R,545,C | NP_004530 | YPRFVQRCTP | YPRFVQRCTP | 18 | 1.59E-05 | 4 | 251022 | RXXP |
| Pathogenic | Collagen type 1 aipha1 | COLIAL | Osteogenesis imperfecta type III | O13 | L,146,P | NP_000079 | GFDVGPVCFP | GFDVGPVCFP | 17 | NILL | NULL | NULL | VXXK |
| Pathogenic | Serpain family C membe | SERPINC 1 | Authrombin III deficincy | AT3D | P,413,L | NP_0013519 | MGRVANLCVK | MGRVANLCVK | 1 | NULL | NULL | NULL | PXXK |
| Pathogenic | tumor suppressorcandi | TUSC3 | mental returdation autosomal resessive 7 | MRT7 | S,343,T | NP_0013433 | HGYPTFLIK | HGYPTFLIK | 8 | NULL | NULL | NULL | SXXXK |
Table 3: Potential pathogenic mutations in key consensus amino acids of new C-terminal minimotifs.
Minimotifs are the carboxyl end of proteins bind to other molecules, traffic proteins to specific sub-cellular compartments, and modify proteins with covalent attachment to other molecules. To explore potential protein interactions in a cellular environment through protein C-termini, we developed an affinity purification LC-MS/MS-based workflow examining new potential C-terminal minimotif sequence patterns. We computationally analyzed the human proteome searching for minimotif patterns with degeneracies that were enriched at the C-termini. Thirty of the putative C-terminal minimotif sequence patterns for approximately 16% of the proteins in the human proteome were selected. These putative C-terminal minimotifs were experimentally tested by affinity LC-MS/MS to identify potential binding partners. We rediscovered 34 previously known interactors in the immunoprecipitates of the 30 putative C-terminal minimotifs. However, while these interactions were previously determined, the region of the interaction was not previously known to bind through the C-terminal minimotif. We also identified 49 potential interactors with matching GO Biological processes for 11 of the 30 predicted C-terminal minimotifs.
We observed a high variance in the number of potential interactors identified in the screen for each of the 30 C-terminal minimotifs ranging from 0 to hundreds. Three of the 30 minimotifs each had 100s of interactors. Thus, about 10% of the novel minimotifs tested may have a more generalizable global role like a PDZ domain interaction minimotif [15,36,37]. We observed that enrichment of proteins having a role in RNA splicing for LxxxI> and DNA damage and cell cycle arrest for QxxL>. This observation implies that there may be many more general C-terminal consensus minimotifs that remain to be discovered. However, we cannot at this point rule out the possibility that the immunoprecipitates in these experiments have a few direct interactions and many indirect interactions as part of larger protein complexes.
Although, the minimotifs studied are not a random sample, approximately 15 of the 30 minimotifs had less than 10 interactors, suggesting that these are functional minimotifs, but may be highly specific. Since this was such a large portion of the C-terminal minimotifs we tested, it suggests that many C-termini may have rather specific minimotifs like the WxxW> minimotif that binds TLE domains [48]. This was not expected because most minimotifs in our C-terminome database have many instances that belong to one consensus sequence [1]. The C-terminome database is likely biased toward high throughput studies since they are easier to annotate from the literature. If this frequency is verified in a larger randomized sample, this result suggests that the C-termini of most proteins have determinants for specific minimotif functions.
One of the 30 minimotifs, RxxR> had no binding partners suggesting that this is not an important minimotif. We also noticed that at least one putative C-terminal minimotif, RxxR>, did not immunoprecipitate any protein and that 15 putative C-terminal minimotifs may have more specific roles such as DEWDx> minimotif in N-WASP binding to Aldolase [49]. However, there are limitations to this screening approach. We tested only one sequence for each pattern, and other instances could produce different results. Alternatively, residues adjacent to the minimotifs or structural context may influence RxxR> interactions.
Given that 30% of the immunoprecipitated proteins had a role in RNA splicing, we wanted to further analyze the profile of the 174 proteins containing LxxxI> at their C-terminus and found that these 174 proteins are endogenously expressed in almost all sub-cellular localizations with varied biological functions. This indicates that the LxxxI> minimotif is not exclusive to nuclear proteins involved in RNA splicing. Although all 10 proteins have at least one serine, threonine and/or tyrosine residue phosphorylated, we did not find the 13 amino acid long recognition minimotif, x[KR]x[KR]x[KR]x[ST]xxRxx, of CLK1 in 10 predicted proteins [44]. This suggests that either LxxxI> minimotif of the CLK1 protein binds with these proteins to form protein complexes for downstream events or there are other unidentified recognition minimotifs of the CLK1 present in these proteins. Alternatively, LxxxI> does not bind to all 10 proteins directly and the co-immunoprecipitated proteins are a part of the large complex.
We identified 1,000s of nonsense variants that causes a premature stop codon and a loss of function of the encoded C-terminal minimotif. With regards to the C-terminome these variants delete the endogenous C-termini and create a new C-terminal sequence. However, nonsense mutations are more difficult to interpret because new C-terminal motifs with new functions may be created, the protein may be expressed at lower levels or not at all, or other functions of C-termini may be deleted. Therefore, we focused on missense mutations in consensus residues for the new minimotifs we identified in this study. Many missense mutants were non-pathogenic or of unknown significance. We expected this result because mutations of large effect tend to be rare and associated with Mendelian disorders.
Therefore, we focused on identifying missense mutations in key residues of the new minimotifs we identified because interpretation was more straight-forward. By designing an informatics search of our new minimotif consensus sequences in the human proteome and the ClinVar database 149 mutations of interest were identified. However, only six where categorized as pathogenic. For example, in mental retardation, TUSC3 has a C-terminal SXXXK minimotif in one isoform that is conserved in mammals and has a conservative Ser to Thr mutation that was noted by the authors not likely to impact structure [50]. However, as our proposed new binding motif, this residue could impact binding, a new hypothesis that could be explored.
Minimotifs on the C-termini are well established as zipcodes for trafficking [51]. Three of the six new mutated C-terminal minimotifs may be involved in trafficking, which would justify why the minimotif is enriched and has multiple instances in the proteome. Mutation of a residue in the SXK minimotif CRYM causes deafness [52]. The mutant CRYM had different subcellular localization as would be expected for a trafficking motif. A G to D mutation that eliminates GXXXL minimotif in CD40LG caused Hyper-IgM syndrome supported by two independent studies [53-55]. In these studies, CD40LG was not detected on the cell surface like wild type protein, suggesting a trafficking deficit. A Leu residue in the VxxL minimotif is mutated to Pro in COL1A1 is mutated and causes Osteogenesis imperfecta type III [56]. This mutation delays collagen triple helix formation and reduced COL1A1secretion, which could be attributed to poor trafficking. These are examples of a systemic approach to identify functional minimotifs on the C-termini and when matched to disease-associated mutations identify new hypotheses for disease mechanisms.
Despite the encouraging results, our workflow has certain limitations that need to be further addressed. One limitation is that alanine substitution mutants used as controls in the affinity experiments is intended to generate loss-of-function minimotifs but may create new minimotif sequences with a recognition for Alanine e.g., AxA>, AxxA> and AxxxA> (Figure S5). These three C-terminal minimotifs containing Ala are present in the human proteome [1] (Figure S6). Although plausible, given that the alanine side chain is less likely to drive protein binding and is commonly used as a negative control. Furthermore, several published interactors were identified in the empty vector control samples. Most of these interactors were identified in a highthroughput manner. One cannot rule out the possibility that those are interactors are indeed false positives or are an artifact of different experimental conditions such as cell lines, affinity beads, epitope tags, the sensitivity of the MS instruments, and computational approaches and parameters for peptide and protein identification. One of the main disappointments in our study, thus far, was the inability to identify a generalizable C-terminal minimotif interaction with protein domain like a PDZ domain-minimotif interaction.
In conclusion, the focus of this research was to explore potential binding partners of the predicted C-terminal minimotif patterns to identify their binding function. The advantages of our approach are the use of small bait peptides narrowing down the interacting region to a specific peptide determinant during the screening process; mammalian expression system and more importantly, the top-down approach of testing the consensus patterns of potential functional degeneracies. Our approach can be used to predict the post-translational modifications of the C-terminal minimotif sequence patterns as well. We have previously cataloged 3,500 C-terminal minimotifs for 17% of the human proteome and characterized the human C-terminome identifying an additional ~9 million repetitive sequence patterns on the C-termini of proteins in the human proteome through computational analysis. In this study, we have identified binders for an additional 1% of the human proteome and inferred functions for an additional 16% of the human proteome. We identify key mutations in the new minimotifs that give insight into potential pathogenic mechanisms for 6 different diseases. Additional validation experiments are needed to confirm interactions and test the role of the new minimotifs in specific disease identified herein. Our proteomics approach has expanded the functional understanding of the entire C-terminome.
MRS conceived, administered, supervised, and obtained funding for the project. SS and MRS conceived, designed, and interpreted experiments. NN, AB, and SS generated all plasmid constructs. SS performed all cell culture and biochemistry experiments related to the affinity LC-MS/MS. XW engineered the program for protein-domain analysis. ZF analyzed the presence of minimotifs in genes and disease. SS and MRS analyzed data and wrote the manuscript.
This work was supported by the National Institutes of Health [R15 GM107983, P20 GM121325, P20 GM103440]; University of Nevada Las Vegas Graduate and Professional Students Association; Prabhu Endowed Professorship; Office of Undergraduate Research, and SIGMA Xi Grants-in-Aid.
All data is included in supplemental files and tables.
We thank the Nevada Proteomics Center (University of Nevada Reno) for proteomics analysis. We thank the MS & Proteomics Resource at Yale University for providing the necessary mass spectrometers and the accompany biotechnology tools funded in part by the Yale School of Medicine and by the Office of The Director, National Institutes of Health (S10OD02365101A1, S10OD019967, and S10OD018034). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank Susan Mockus (Cedars Sinai Hospital) for suggestions in editing the manuscript.
Martin Schiller is a CEO of Heligenics, Biotechnology Company, but the contents of this paper are not related to the business. The other authors have no competing interests to declare.
Citation: Sharma S, Novakovic N, FitzHugh ZT, Bragg A, Brooks S, Wu X, et al. (2021) New Minimotifs and their functions in the C-Terminome. J Proteomics Bioinform.14:561.
Received: 13-Dec-2021 Accepted: 27-Dec-2021 Published: 03-Jan-2022
Copyright: © 2021 Sharma S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.