Research Article - (2015) Volume 1, Issue 1
Microcosm Assessment of the Effect of an Acute Mercury Contamination Event on the Structure and Activity of Sediment Bacterial Communities
*Corresponding Author(s): Ana I Lillebo, Department of Biology and CESAM, University of Aveiro, Campus de Santiago, 3810-193 Aveiro, Portugal, Tel: +351-234-370-779, Fax: +351-234-426 408 Email: Angela Cunha, Department of Biology and CESAM, University of Aveiro, Campus de Santiago, 3810-193 Aveiro, Portugal, Tel: 351-234-370-784, Fax: +351-234-426 408 Email:
Abstract
Objectives: In the present study, the effect of acute mercury contamination on the structure and activity of bacterial communities in intertidal mudflats was assessed through a microcosm experiment simulating the mobilization of highly contaminated sub-surface sediments.
Methods: Box-microcosms corresponding to different test conditions were constructed by mixing natural estuarine sediments with high and low concentrations of mercury in defined proportions. The effects on sediment bacteria were characterized by quantifying bacteria using fluorescent in situ hybridization (FISH) technique, assessing the community structural diversity by denaturating gradient gel electrophoresis (DGGE) and analysing descriptors of bacterial activity (extracellular enzymatic activity and leucine incorporation) at the beginning and at the end of a 7-days incubation period.
Results: At the end of the experiment, total abundance of Bacteria was significantly higher in the low-Hg microcosms than in the high-Hg and blended-sediment microcosms. DGGE patterns revealed that the structure of sediment bacterial communities responded to the experimental treatment and to the incubation time. Bacterial activity was inhibited by mercury and that the levels of arylsulfatase and biomass productivity were inversely related with the Hg concentration. The proportion sulfate-reducing in relation to total prokaryotes increased at the end of the experiment, which might indicate a differential response of Bacteria and Archaea to confinement and mercury contamination.
Conclusion: Mechanical disturbance of sediments historically exposed to mercury contamination, like bottom trawling or dredging, will cause the mobilization of deeper sediments highly contaminated with Hg which will impact the less contaminated surface sediments. These acute events will impact the structure and activity of bacterial communities and their contributions to the associated biogeochemical cycles, with expectable impacts at the ecosystem level.
Keywords: Mercury; Bacterial communities; Sediment resuspension; Sulfate-reducing bacteria; Bacterial activity; Acute toxicity; Estuary
Introduction
Mercury is a highly toxic element that may be present in aquatic environments as a consequence of anthropogenic activities. By being persistent, easily dispersible and susceptible of bioaccumulations, mercury is considered a global concern pollutant [1]. Although the severe environmental regulations implemented in the last decades have restrained anthropogenic emissions, mercury accumulated in sediments, known as historical contamination, is still a threat to the health of aquatic ecosystems because of the potential release to other environmental compartments (atmosphere, water column, living organism) [2].
Salt marshes are particular environments with a worldwide recognized importance in providing essential ecological functions and services [3,4]. However, as transitional areas, they are more susceptible to large inputs of pollutants, namely mercury, mainly from anthropogenic sources [5,6]. Integrated interventions to minimize the effects of historical contamination by mercury have been set up in coastal ecosystems and wetlands around the world [7-9]. Recent studies have attempted to understand the mechanisms of mercury storage in salt mash sediments and the role of plants in mercury sequestration and decontamination [10,11]. Long term exposure to mercury is known to cause changes in structure and function of sediment bacterial communities [12-16] and the methylation of mercury to the neurotoxic species methyl mercury (CH3Hg+) is directly related to the heterotrophic activity of anaerobic bacteria. Although iron-reducing bacteria [17] and methanogens [18] can contribute to mercury methylation, and the recent evidence of this capacity in novel bacterial and archaeal strains [19], sulfate-reducing bacteria (SRB) are still regarded as major players in methylmercury production [20-22]. SRB belonging to the family Desulfobacteriaceae (Deltaproteobacteria class) that use acetate as electron donor are recognized as active mercury methylators [20,23].
The Laranjo basin, a 2 km2 inner area of the Ria de Aveiro coastal lagoon, was exposed to intense contamination with mercury during the second half of the XX century. Although the industrial processes that caused this situation have changed, and mercury levels in the water column and surface sediments are in now in compliance with environmental regulations, large amounts of mercury are still present in the sub-surface sediments [7,24]. Due to human activities such as bottom trawling or dredging, or because of physical, chemical and biological processes, such as hydrodynamic flows, bioturbation or chemical transformation, Hg can be remobilized [9,25,26]. The mobilization of mercury accumulated in deeper sediments will affect the distribution of mercury within the sediment column, the processes of metal speciation and the transfer between the sediment compartment and the water column [26-29].
The aims of these experiments were: (i) to infer on the impact of a possible dredging on the structure and activity of sediment bacterial communities and (ii) to quantify the sulfate-reduction bacteria, as likely players in processes of Hg methylation. In order to simulate the mobilization of highly contaminated sediments from sub-surface sediments by dredging or other form of mechanical disturbance, control-sediment (low concentration of Hg) from the surface were experimentally mixed with deeper sediments (0.4-0.5 m) from the corresponding site (high concentration of Hg) in a microcosm experiment.
Materials and Methods
Study area
The Ria de Aveiro is a temperate shallow coastal lagoon located at Northwest Atlantic coast of Portugal (40º38’N, 8º 44’W). This mesotidal system forms a complex network of channels with extensive intertidal mudflats that get exposed during low tide. In this system, the small inner Laranjo basin is historically contaminated with mercury discharged from a mercury cell chlor-alkali plant located in Estarreja industrial complex and presently, deeper sediments (down to 0.4-0.5 m) show the highest contamination levels [7]. Having in mind the well recorded history of the system, sediment samples were collected at low tide from an unvegetated area adjacent to the salt marsh in the Laranjo Basin. Sediment horizons corresponding to the 0-0.05 cm (surface) and 0.4-0.5 m (deeper) were collected in the field, placed in sterile plastic bags, and kept in the cold during transport to the laboratory.
Sediment characterization
In the laboratory, each sediment layer was homogenized and cleaned from shells and other debris. Homogenised sub-samples were analysed for the percentage of fine particles (<0.63 mm), and total mercury content (ng Hg g-1). The total mercury concentration in sediments from 0-0.05 and the 0.4-0.5 m sediment horizons was directly determined by atomic absorption spectrometry following thermal decomposition with an Advanced Mercury Analyser (AMA) LECO 254 (Costley et al. 2000). In order to assess the accuracy and precision of the analytical methodology, analysis of certified reference material was carried out (PACS-2 harbour sediment) in parallel with samples [24]. Certified and measured values were in agreement with recoveries of 95% for PACS 2. Mercury quantifications were performed in triplicate and blanks were run in parallel.
Experimental set-up
In the laboratory, the homogenised sediment was distributed in microcosm (9.5 × 9.5 × 4 cm acid-cleaned PET). The test condition (blended 1:4) was achieved by mixing the positive (high-Hg) and the negative (low-Hg) control sediments in a proportion of 1:4 (w:w). Each box-microcosm contained a total of 200 g of sediment and four replicate box-microcosms were prepared for each experimental condition. Microcosms were incubated at room temperature (22°C), at natural light. The biological effects of acute mercury contamination on sediment bacterial communities were assessed by analysing the community structure and descriptors of bacterial activity at the beginning and at the end of the incubation (7 days).
Extracellular arylsulfatase activity
The activity of arylsulfatase was determined fluorimetrically using a Jasco FP-777 fluorometer (Jasco Inc. Easton, MD USA) in sediment suspensions [30]. MUF-sulfate (Sigma-Aldrich®, St. Louis MO, USA) was as used as a fluorogenic labelled substrate. The final saturating concentration, established by preliminary kinetic assays, was 2 mM. Sediment suspensions were prepared by adding 100 ml of sterile diluted artificial seawater (salinity 17, same as salinity in situ) to 1 g of fresh sediment and stirred in order to obtain homogeneous sediment suspensions. Six aliquots were transferred to 2 ml microtubes and added of the stock substrate solution. The initial fluorescence (λext=365 nm and λem=450 nm) was read in three of replicates, after centrifugation (13,000 × g, 5 min) for the removal of particles, and alkalinisation with 100 μl of a buffer solution (1.384 ml of ammonium, 0.375 g glycin and distilled water to 100 ml, pH 10.5) in order to enhance MUF fluorescence. The remaining three aliquots were incubated at room temperature (22°C) for 3 hours, after which particles were removed by centrifugation, the buffer solution was added and the final fluorescence was read. The rate of substrate hydrolysis was estimated from the variation of fluorescence, standardized to 1 hour incubation and converted to units of substrate concentration with a calibration curve prepared for the fluorescence product (MUF) by the internal standard approach.
3H-leucine incorporation
3H-Leucine incorporation [31] was used to estimate bacterial biomass productivity (BBP). For each sample, three aliquots of sediment suspension plus one trichloroacetic acid (TCA)-killed control were placed into Eppendorf tubes and incubated with 483 nM of 3Hleucine (Amersham, Biosciences Ltd, Sweden, 45-80 Ci/mmol) for 1 h at room temperature. The saturation concentration and the incubation time were established in preliminary kinetics assays. Incubation was stopped with 80 μl of TCA (100%). Samples were centrifuged (13.000 g for 10 min), the supernatant was rejected, and 1 ml of cold TCA (5%) was added. Between centrifugations, 1 ml of cold TCA (5%) and cold ethanol was added. Finally, 1 ml of scintillation cocktail was added. After a 3 day period of stabilization, the radioactivity was determined in a scintillation counter (Beckman LS 6000 IC; GMI, Inc, Ramsey, Minnesota, USA).
Quantification of bacteria and sulfate reducing bacteria (SRB)
Quantification of Bacteria and SRB was performed by Fluorescent In Situ Hibridization (FISH) with a mixture of the oligonucleotide probes EUB338 [34], EUB338 II and EUB338 III (Daims et al. 1999) for the Bacteria domain and a mixture of the oligonucleotide probes DELTA 495 a, b e c, covering most Deltaproteobacteria for sulfate reducing bacteria [32,33]. The protocol was adapted from Llobet- Brossa et al. [34], with minor modifications. Samples (0.5 g of fresh sediment) were fixed in 2% formaldehyde. Fixed samples were washed with 1 × PBS, centrifuged (13000 × g for 2 min) and stored in PBS/ ethanol (1:1) at -20ºC. Aliquots of sediment suspension were diluted with PBS, mixed, and cells were collected by filtration on the surface of 0.2 μm pore-size polycarbonate membranes (GE Osmonics Labstore, Minnetonka, USA) [35]. Hybridizations were performed with 21 μl of hybridization buffer (900 mM NaCl, 20 mM Tris/HCl, pH 8, 0.01% SDS) for both Bacteria and SRB probes (Eurofins MWG Operon, Ebersberg, Germany) and 3 μl of each probe (5 ng/μl), at 46ºC for 90 min. After hybridization, the membranes were washed with a buffer solution for 15 min at 48ºC. The membranes were subsequently rinsed with distilled water, air-dried, stained with DAPI, mounted in Citifluor immersion oil solution (Citifluor Ltd, London, UK) and examined under an epifluorescence microscope equipped with a mercury bulb and filter sets 31000 (Chroma) for DAPI detection and 41007a (Chroma) for Cy3 detection. Cells were counted with 1000 × magnification in 10 randomly selected optical fields. Total DAPI counts were an estimate of total number of prokaryotic cells [35].
16S rRNA denaturing gradient gel electrophoresis (DGGE) profiling of the bacterial communities
DNA was extracted from sediment using the E.Z.N.A. Soil DNA kit (Omega Bio-Tek, Norcross, GA, USA) according to the instruction of the manufacturer. PCR amplification was carried out in a Veriti 96- Well Thermal Cycler (Applied Biosystems, Foster City, CA, USA) using DreamTaq™ PCR Master Mix (2X) purchased from Fermentas (Fermentas Canada Inc., Burlington Ontario, Canada) and nucleotides from IBA GmbH (IBA GmbH, Göttingen, Germany). 16S rRNA gene fragments were amplified with general bacterial primers U27F (5´- AGAGTTTGATCCTGGCTCAG-3´) and 1494L (5´- GGTTACCTTGTTACGACTT-3´) [36]. Reaction mixtures (25 μl) contained 1 μl of bovine serum albumin (2 mg ml-1, diluted 100 x), 0.1 μM of each DNA primer, 12.5 μl HotStartTaq™ PCR Master Mix (Qiagen, Venlo, The Netherlands) and 1 μl template DNA. The amplification conditions were as follows: initial denaturation (95ºC for 10 min); 25 cycles of denaturation (95ºC for 45 s), annealing (56ºC for 45 s), extension (72ºC for 1:30 min); final extension (72ºC for 10 min). For all samples, a nested PCR procedure was followed. The primers for the amplification were 685 GC (5´-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAA CGC GAA GAA CCT TAC-3´) and 1401L (5´-CGG TGT GTA CAA GAC CC-3´) [37] and a GC clamp was attached to the 5´end of the forward primer to increase the separation of DGGE bands in the electrophoretic analysis. Each reaction (25 μl) contained 0.5 μl of acetamide (50%), 0.1 μM of each DNA primer, 12.5 μl HotStartTaq™ PCR Master Mix and 1 μl of template DNA. The amplification protocol was: an initial denaturation at 95ºC for 10 min, 30 cycles of denaturation at 95ºC for 1 min, annealing at 53ºC for 1 min, extension at 72ºC for 1:30 min, final extension at 72ºC for 7 min, and cooling to 15ºC. Negative controls without template DNA were included. Five μl of PCR products were analysed by electrophoresis on a 1.5% agarose gel with GelRed (Biotium, Hayward, CA, USA) as DNA staining agent.
DGGE was performed using the DCodeTM Universal Mutation Detection System (Bio Rad Laboratories, Hercules, CA, USA). PCR amplicons were loaded onto 6-10% (w/v) polyacrylamide gel in 0.5 × TAE buffer (20 mM Tris-acetate pH 7.4, 10 mM sodium acetate, 0.5 mM Na2EDTA) using a linear denaturing gradient ranging from 40% to 58%. Electrophoresis was performed for 16 h at 80 V at 60°C in 1 x TAE buffer. The gels were silver-stained [38]. The images were analysed using the software package Bionumerics (Applied Maths, Sint- Martens-Latem, Belgium).
Statistics
The significance of the differences between treatments was assessed by 2-way ANOVA using the SPSS Statistics 17.0 (International Business Machine Corp., New York, USA). Normality was confirmed by Kolmogorov-Smirnov test and equality of variances was checked by the Levene test.
The data matrix of band (band positions and their corresponding intensities) abundance per sample was log10 (x + 1) transformed and a distance similarities matrix constructed using the Bray-Curtis index with the vegdist function in the vegan package in R version 2.11.1 [39]. Variation in bacterial composition among treatments/time was visually assessed with Principal Coordinates Analysis (PCO) using the cmdscale function in R and the Bray–Curtis distance matrix as input. Variation among treatments groups was compared using the Adonis function in vegan. The Adonis function is an analysis of variance with distance matrices using permutations that partitions distance matrices among sources of variation; in this case treatment and time. In the adonis analysis, the Bray-Curtis distance matrix of band composition was the response variable with treatment (high-Hg, low-Hg and blended 1:4) and time (day: 0 or 7) as independent variables. We also included the interaction term between treatment and time. The number of permutations was set at 999; all other arguments used the default values set in the function. Measured microbiological variables, namely biomass productivity (μgC.gdw-1h-1), arylsulfatase (nmol.gdw-1h-1), number of prokaryotic cells (cells.gdw-1), number of bacteria (cells.gdw-1), number of sulfate reducing bacteria (cells.gdw-1), proportion of EUB (EUBP: EUB abundance/ total prokaryote abundance (NTP)), proportion of SRB (SRBP: SRB abundance/ total prokaryote abundance (NTP)) and Hg content (ngHg.mgdw-1) were fit in the PCO ordinations using the envfit function in vegan. The envfit function was also used to test for significant relations between these variables and the PCO ordinations of bacterial community composition using 999 permutations; all other arguments in the function were left as default.
Results
Sediment characterization
The surface sediment (0-0.05 m), used as low-Hg, was characterized by 60.2 ± 3.3% of fine particles (<0.63 mm) and 14.0 ± 0.1 ngHg.mgdw-1. In the deeper high- Hg sediments (0.4-0.5 m) used as the source of mercury for the microcosm experiments, the percentage of fine particles was 59.2 ± 1.5% and the concentration of mercury was 98.9 ± 1.7 ngHg.mgdw-1.
Extracellular enzymatic activity
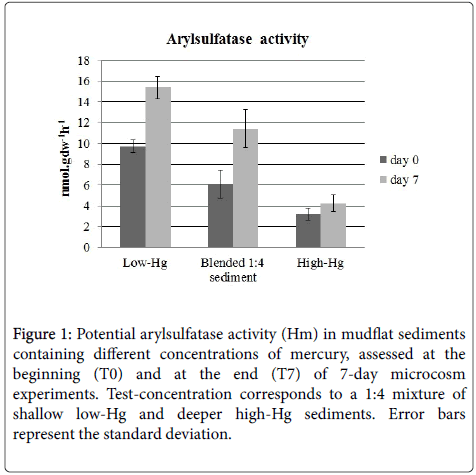
The variation of arylsulfatase activity in the conditions of the experiments is represented in Figure 1. At the beginning of the experiment, arylsulfatase activity was the highest in samples from the upper sediment layer, taken as low-Hg (9.7 nmol.gdw-1h-1), and lowest in sediments from the 0.4-0.5 m, taken as the high-Hg (3.2 nmol.gdw-1h-1). In blended 1:4 sediment, the potential maximum hydrolysis rate (Hm) was intermediate (6.1 nmol gdw-1h-1) between both control sediments and differences between treatments were statistically significant (two-way ANOVA, p<0.05).
Figure 1: Potential arylsulfatase activity (Hm) in mudflat sediments containing different concentrations of mercury, assessed at the beginning (T0) and at the end (T7) of 7-day microcosm experiments. Test-concentration corresponds to a 1:4 mixture of shallow low-Hg and deeper high-Hg sediments. Error bars represent the standard deviation.
At the end of the experiment, the pattern of variation was similar but there was a statistically significant overall increase (two-way ANOVA, p<0.05) in activity in the low-Hg sediment (15.4 nmol.gdw-1h-1) and in blended 1:4 (11.4 nmol.gdw-1h-1), in relation to the beginning of the incubation. At the end of the experiment, arylsulfatase Hm in the high-Hg microcosms (4.22 nmol.gdw-1h-1) was not significantly different from the initial value.
Bacterial biomass productivity
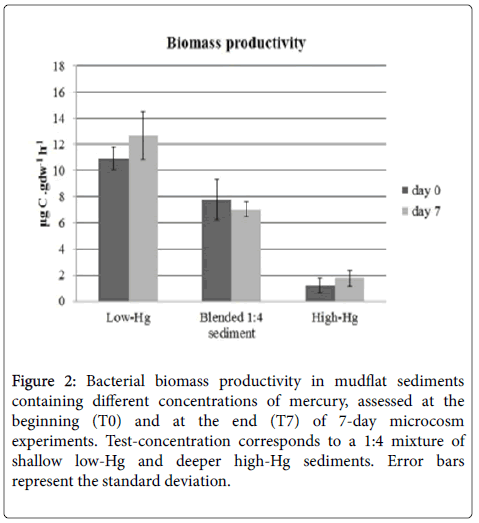
The variation of the rate of bacterial biomass productivity is represented in Figure 2. The highest activity rate was observed in the low-Hg (10.9 μgC.gdw-1h-1) and the lowest in the high-Hg (1.2 μgC.gdw-1h-1). In blended 1:4 sediment, biomass productivity was 7.8 μgC.gdw-1h-1. Differences between treatments were statistically significant (two-way ANOVA, p < 0.05) but differences between the beginning and the end of the microcosm experiments were not.
Figure 2: Bacterial biomass productivity in mudflat sediments containing different concentrations of mercury, assessed at the beginning (T0) and at the end (T7) of 7-day microcosm experiments. Test-concentration corresponds to a 1:4 mixture of shallow low-Hg and deeper high-Hg sediments. Error bars represent the standard deviation.
Prokaryote abundance
Total number of prokaryotes was the highest (2.2 ± 0.22 × 109 cells.gdw-1) in low-Hg, the lowest (1.0 ± 0.06 × 109 cells.gdw-1) in high-Hg and intermediate (1.3 ± 0.06 × 109 cells.gdw-1) in the blended 1:4 sediments (Table 1). During the 7-day incubation, the variation in total prokaryote abundance was not statistically significant in any of the treatments (Table 1).
| Samples | Absolute abundance (x 109 cells.gdw-1) | Relative abundance (%) | ||||
|---|---|---|---|---|---|---|
| TPN1 | Bacteria | SRB2 | Bacteria/TPN | SRB/TPN | ||
| Low-Hg | T0 | 2.2 ± 0.22 | 1.9 ± 0.21 | 0.5 ± 0.11 | 82.8 ± 1.44 | 23.8 ± 4.08 |
| Blended 1:4 sediment | 1.3 ± 0.06 | 0.9 ± 0.09 | 0.4 ± 0.02 | 69.4 ± 5.31 | 32.6 ± 0.72 | |
| High-Hg | 1.0 ± 0.06 | 0.5 ± 0.04 | 0.2 ± 0.02 | 53.9 ± 5.93 | 20.0 ± 1.88 | |
| Low-Hg | T7 | 2.6 ± 0.16 | 2.0 ± 0.22 | 1.1 ± 0.22 | 76.2 ± 4.40 | 40.2 ± 7.39 |
| Blended 1:4 sediment | 1.4 ± 0.08 | 1.0 ± 0.05 | 0.5 ± 0.04 | 73.1 ± 6.71 | 33.7 ± 4.19 | |
| High-Hg | 1.0 ± 0.06 | 0.7 ± 0.03 | 0.4 ± 0.10 | 62.6 ± 0.98 | 39.1 ± 10.18 | |
TPN: Total number prokaryote; SRB: sulfate reducing bacteria
Table 1: Abundance of prokaryote cells (Bacteria and SRB) ± SD (4 replicates) in mudflat sediments containing different concentrations of mercury, assessed at the beginning (T0) and at the end (T7) of 7-day microcosm experiments.
On average, Bacteria detected with the probes EUB338, EUB338II or EUB 338III accounted for 70% DAPI stained cells (total prokaryotes). At the beginning of the experiment, the abundance of Bacteria was significantly higher (two-way ANOVA, p<0.001) in low- Hg (1.9 ± 0.21 × 109 cells.gdw-1, 82.8 ± 1.44% TPN), than in high-Hg or blended 1:4 sediments. After 7 days of incubation, the values of total abundance of Bacteria detected with the probes EUB338, EUB338II or EUB 338III were not significantly different from the initial values, with exception of high-Hg (ANOVA, p<0.05). The abundance of Bacteria was still significantly higher in the low-Hg (2.0 ± 0.22 × 109 cells.gdw-1, 76.2 ± 4.40% TPN) and lower in blended 1:4 sediments (1.0 ± 0.05 × 109 cells.gdw-1, 73.1 ± 6.71% TPN) and in the high-Hg (0.7 ± 0.03 × 109 cells.gdw-1, 62.6 ± 0.98% TPN).
SRB accounted for 30% of total prokaryote abundance, on average. At the beginning of the experiment, the proportion of SRB in relation to total prokaryotes was higher in low-Hg sediments than in subsurface sediments. During the experiment, there was a significant increase in the proportion of SRB in all samples, with exception of blended 1:4 sediment (two-way ANOVA, p<0.001). At the beginning of the experiment, the abundance of SRB in low-Hg (0.5 ± 0.11 × 109 cells.gdw-1; 23.8 ± 4.08% TPN) and blended 1:4 sediment (0.4 ± 0.02 × 109 cells.gdw-1; 32.6 ± 0.72% TPN) was significantly higher than in the high-Hg (0.2 ± 0.02 × 109 cells.gdw-1; 20.0 ± 1.88% TPN) (two-way ANOVA, p<0.001). During the 7-day incubation this pattern persisted but there was an increase in SRB abundance (Table 1).
Community structure
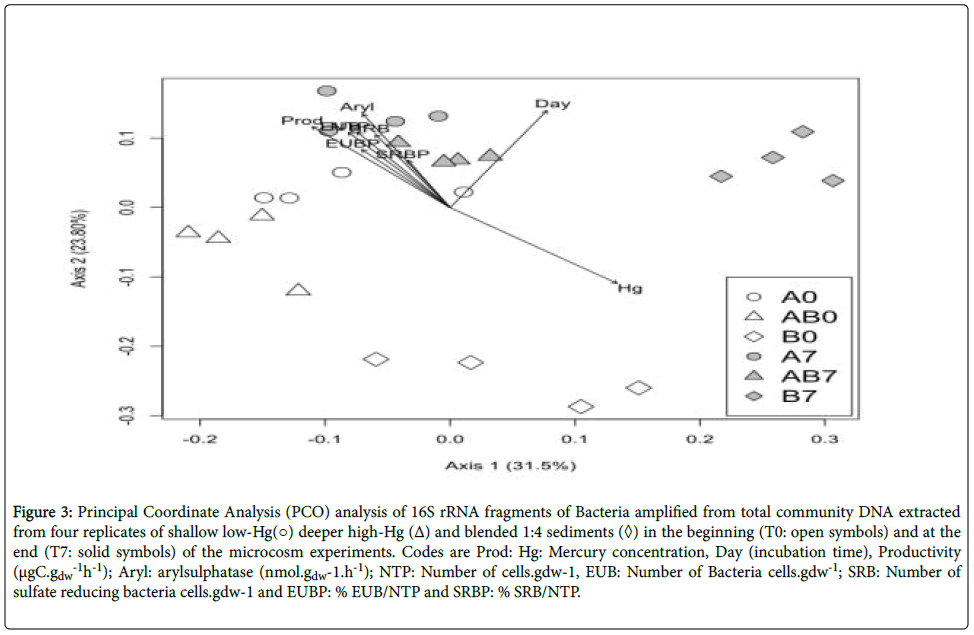
DGGE profiles bacterial 16S rRNA gene fragments revealed differences in the banding pattern between the high-Hg and the other two sediments treatments (low-Hg and blended 1:4 sediments) related with treatment (Adonis: F2,23=5.080, R2=0.326, P<0.001), time (Adonis: F1,23=6.223, R2=0.221, P<0.001) and the interaction between treatment and time (Adonis: F5,23=9.060, R2=0.716, P<0.001). PCO ordination of DGGE profiles is represented in Figure 3. The first two PCO axes explain approximately 55% of the variability of the dataset. Generally, replicates from the same experimental treatment group closely, indicating a good reproducibility of the analysis.
Figure 3:Principal Coordinate Analysis (PCO) analysis of 16S rRNA fragments of Bacteria amplified from total community DNA extracted from four replicates of shallow low-Hg(○) deeper high-Hg (Δ) and blended 1:4 sediments (◊) in the beginning (T0: open symbols) and at the end (T7: solid symbols) of the microcosm experiments. Codes are Prod: Hg: Mercury concentration, Day (incubation time), Productivity (μgC.gdw -1h-1); Aryl: arylsulphatase (nmol.gdw-1.h-1); NTP: Number of cells.gdw-1, EUB: Number of Bacteria cells.gdw-1; SRB: Number of sulfate reducing bacteria cells.gdw-1 and EUBP: % EUB/NTP and SRBP: % SRB/NTP.
The first PCO axis (Figure. 3) reveals that low-Hg and blended 1:4 sediments were characterized by higher arylsulfatase (Aryl; envfit for 1st and 2nd axes: P=0.001), productivity (Prod; envfit for 1st and 2nd axes: P=0.001), total prokaryote abundance (NTP; envfit for 1st and 2nd axes: P=0.001), abundance of Bacteria (EUB; envfit for 1st and 2nd axes: P=0.001), abundance of sulfate-reducing bacteria (SRB; envfit for 1st and 2nd axes: P=0.004) and proportion of Bacteria relation to total prokaryote abundance (EUBP; envfit for 1st and 2nd axes: P=0.012). The high-Hg treatment was characterize by a significant dependence on Hg content and time (Hg and Time; envfit for 1st and 2nd axes: P=0.004). There were no significant associations between the proportion of sulfate-reducing bacteria in relation to total prokaryote abundance (SRBP) and the ordination of the 1st and 2nd axes.
Discussion
A microcosm approach, in which a mechanical disturbance was simulated by mixing sediments from the deeper highly contaminated and surface less-contaminated layers, was carried as a realistic approach to investigate the impact of a possible dredging on the structure and activity of bacterial communities, without the use of an exogenous mercury source.
Hetetrotrophic activity
The descriptors of activity analysed in this work show that bacterial activity was inhibited by mercury contamination and that the levels of arylsulfatase and biomass productivity were inversely related with the concentration of mercury in the sediments. One of the effects of mercury is the inhibition of bacterial heterotrophic activity [40]. Therefore, mercury acute contamination may have an immediate impact on organic matter diagenesis in estuarine sediments.
The bacterial community in the blended 1:4 sediments displayed levels of activity that were intermediate between low-Hg and high-Hg sediments. However, the levels of activity observed in in blended 1:4 sediments cannot be fully explained by processes of conservative mixing of two original communities expressing high (low-Hg) and low (high-Hg) levels of activity. In fact, heterotrophic activity in the blended 1:4 sediments was below the expected (e.g., lower that the weighted average of the values of activity in the two contrasting sediment types), possible indicating that an inhibition at cellular level of the more active community of the upper layer sediments. The comparison of the values observed and those predicted by conservative mixture indicate that the inhibition was immediate (observed in T0) and the inhibition factors were of 28% for arylsulfatase and 13% for biomass productivity. During the incubation, there was a slight recovery of arylsulfatase (13% inhibition at the end of the experiment) but an intensification of the inhibition of biomass production (33% at the end of the experiment). The different pattern of variation of these two descriptors of heterotrophic activity is probably related with the organisms involved. Leucine incorporation is a direct measure of prokaryote biomass productivity, because only prokaryotes are able to take up leucine in the dissolved form. On the contrary, arylsulfatase, although mainly originating from bacteria, is also common in fungi and can be expressed in plants and animals [41] and can be used as an indicator of recovery of contaminated soils [42]. Considering that in sediments, organic matter is mostly available as polymers, extracellular enzymatic activity is the initial and limiting step in organic matter decomposition and recycling [43]. In estuarine and coastal sediments, organic sulfur corresponds to a major fraction of the available sulfur and arylsulfatase activity significantly contributes to sulfate supply [44] for assimilatory or dissimilatory (sulfate reduction) metabolic pathways. In addition, arylsulfatase in soils in can be truly extracellular, meaning that a significant fraction may occur adsorbed to sediment particles [45]. Therefore, the inhibition of arylsulfatase activity may be a combined process of direct inhibition of the enzyme and decreased activity of the organisms producing it, some of which may resist and recover [40]. The decrease in bacterial biomass productivity indicates a decrease in prokaryote activity rate from which cell did failed to recover within the time-frame of the experiment.
Community structure
Communities of Bacteria in the deeper (high-Hg) and surface sediments (low-Hg) were structurally distinct, such as expected from a different degree of exposure to mercury [13] and different environmental conditions, namely sediment texture, availability or organic substrates and electron acceptors [46,47]. Also, the influence of plant roots may have contributed to the in situ shaping of the community structure [10]. In sediments chronically contaminated with mercury, there is a selective pressure towards the increase of the relative abundance of bacterial groups that are tolerant to the metal [15], capable of extracellular bioaccumulation [48] or that are, in some way, involved in processes of speciation and transference, such as sulfate-reducing bacteria [14,49,50]. However, in this work, the proportion of SRB in relation to total prokaryotes in the initial surface sediments (low-Hg) was higher than in deeper sediments (high-Hg). Changes in the structure of sediment bacterial communities during microcosm experiments are likely to [51]. The increase in the proportion of SRB in all samples at the end of the experiment is more likely to have been caused by sediment handling, confinement and oxygen depletion, which may have selected for anaerobes, than an effect of mercury contamination, since it occurred in control and test sediments. The decrease in the proportion of Bacteria, in relation to total prokaryotes, in the positive control and in test sediments, and the opposite trend in the negative control, may indicate a differential response of Bacteria and Archaea to confinement and mercury contamination. Significant effects of mercury contamination on archaeal sediment communities have been demonstrated [52]. In this study, Archaea seem to have been even more responsive than Bacteria, in terms of abundance.
At the beginning of the experiment, the communities in low-Hg, high-Hg and blended 1:4 sediments were different between each other. However, low-Hg and blended 1:4 sediments were more similar between each other than with the communities in the heavily contaminated sediments. Community structure in high-Hg changed during the experiment but at the end of the incubation, it was still different from all the other communities, indicating that the effect of mercury on the shaping of the community structure was stronger than other effects related with confinement or oxygen depletion. The structure of the community in the low-Hg sediment was more responsive to the incubation time and changed in a scattered way. Overall, communities in sediments with the lowest mercury load became different from the initial community in these sediments and more similar to the blended 1:4 community. The structure of the blended 1:4 community was the least variable during the incubation. Taking into consideration that the effects of mercury on bacterial activity were immediate, as inferred from the rates of arylsulfatase activity and leucine incorporation in the blended 1:4 sediments at the beginning of the experiment, community structure seems to be much less reactive to mercury than heterotrophic activity. The changes in community structure observed during the experiment are, therefore, more likely triggered by factors associated with confinement, to which the low-Hg (less subjected to the selective pressure of mercury) was more responsive.
Conclusion
The results of this work demonstrate that the contamination of sediments with realistic concentrations of mercury has a higher impact on bacterial activity than on the structure of bacterial communities. Events causing disturbance or re-suspension of contaminated sediments susceptible of mobilizing mercury accumulating in deeper layers to upper or surface layers will impact bacterial communities and therefore their contributions to the associated biogeochemical cycles. Ultimately, when associated to the re-suspension of sediment particles to the water column, this might have an impact at the ecosystem level.
Acknowledgements
This work was supported by the Centre for Environmental and Marine Studies, University of Aveiro (FCOMP-01-0124- FEDER-007378, Pest C/MAR/LA0017/2011). The Portuguese Foundation for Science and Technology (FCT) supported this study through the project PTDC/MAR/67752/2006, PEst-C/MAR/ LA0017/2013. Financial support to V. Oliveira was provided by FCT in the form of a PhD grant (SFRH/BD/46977/ 2008).
References
- Driscoll CT, Mason RP, Chan HM, Jacob DJ, Pirrone N (2013) Mercury as a global pollutant: sources, pathways, and effects. Environ Sci Technol 47: 4967-4983.
- Lillebø AI, Coelho PJ, Pato P, Válega M, Margalho R, et al. (2011) Assessment of mercury in water, sediments and biota of a southern European estuary (Sado estuary, Portugal). Water Air Soil Poll 214: 667-680.
- Richardson CJ (1999) Plenary session presentation: ecological functions of wetlands in the landscape. Ecotoxicology and risk assessment for wetlands: Pensacola, Florida, Society of Environmental Toxicology and Chemistry (SETAC).
- Barbier EB, Hacker SD, Kennedy C, Koch EW, Stier AC, et al. (2011). The value of estuarine and coastal ecosystem services. Ecol Monogr 81: 169-193.
- Xu Y, Sun Q, Yi L, Yin X, Wang A, et al. (2014) The source of natural and anthropogenic heavy metals in the sediments of the Minjiang River Estuary (SE China): Implications for historical pollution. Sci Total Environ 493: 729-736.
- Wetzel MA, Wahrendorf DS, Peter C (2013) Sediment pollution in the Elbe estuary and its potential toxicity at different trophic levels. Sci Total Environ 449: 199-207.
- Pereira M, Lillebø A, Pato P, Válega M, Coelho J, et al. (2009) Mercury pollution in Ria de Aveiro (Portugal): a review of the system assessment. Environ Monit Assess 155: 39-49.
- Henriques B, Rocha LS, Lopes CB, Figueira P, Monteiro RJ, et al. (2015) Study on bioaccumulation and biosorption of mercury by living marine macroalgae: Prospecting for a new remediation biotechnology applied to saline waters. Chem Eng J 281: 759-770.
- Randall PM, Chattopadhyay S (2013) Mercury contaminated sediment sites - An evaluation of remedial options. Environ Res 125: 131-149.
- Cleary DFR, Oliveira V, Gomes NCM, Pereira A, Henriques I, Marques B, et al. (2012) Impact of sampling depth and plant species on local environmental conditions, microbiological parameters and bacterial composition in a mercury contaminated salt marsh. Mar Pollut Bull 64: 263-271.
- Figueiredo NL, Areias A, Mendes R, Canário J, Duarte A, et al. (2014) Mercury-resistant bacteria from salt marsh of tagus estuary: the influence of plants presence and mercury contamination levels. J Toxicol Env Heal A 77: 959-971.
- Poulain AJ, Aris-Brosou S, Blais JM, Brazeau M, Keller W, et al. (2015) Microbial DNA records historical delivery of anthropogenic mercury. ISME J
- Vishnivetskaya TA, Mosher JJ, Palumbo AV, Yang ZK, Podar M, et al. (2011) Mercury and other heavy metals influence bacterial community structure in contaminated Tennessee streams. Appl Environ Microbiol 77: 302-311.
- Macalady JL, Mack EE, Nelson DC, Scow KM (2000) Sediment Microbial Community Structure and Mercury Methylation in Mercury-Polluted Clear Lake, California. Appl Environ Microbiol 66: 1479-1488.
- Rasmussen LD, Sørensen SJ (1998) The effect of longterm exposure to mercury on the bacterial community in marine sediment. Curr Microbiol 36: 291-297.
- Rieder SR, Frey B (2013) Methyl-mercury affects microbial activity and biomass, bacterial community structure but rarely the fungal community structure. Soil Biol Biochem 641: 64-173.
- Kerin EJ, Gilmour C, Roden E, Suzuki M, Coates J, et al. (2006) Mercury methylation by dissimilatory iron-reducing bacteria. Appl Environ Microbiol 72: 7919-7921.
- Hamelin S, Amyot M, Barkay T, Wang Y, Planas D (2011) Methanogens: principal methylators of mercury in lake periphyton. Environ Sci Technol 45: 7693-7700.
- Gilmour CC, Podar M, Bullock AL, Graham AM, Brown SD, et al. (2013) Mercury methylation by novel microorganisms from new environments. Environ Sci Technol 47: 11810-11820.
- Barkay T, Wagner-Dobler I (2005) Microbial transformations of mercury: potentials, challenges, and achievements in controlling mercury toxicity in the environment. Adv Appl Microbiol 57:1-52.
- Hines ME, Evans RS, Sharak Genthner BR, Willis SG, Friedman S, et al. (1999) Molecular phylogenetic and biogeochemical studies of sulfate-reducing bacteria in the rhizosphere of Spartina alterniflora. Appl Environ Microbiol 65: 2209-2216.
- Liu YR, Yu RQ, Zheng YM, He JZ (2014) Analysis of the microbial community structure by monitoring an Hg methylation gene (hgcA) in paddy soils along an Hg gradient. Appl Environ Microbiol 80: 2874-2879.
- Yu RQ, Flanders J, Mack EE, Turner R, Mirza MB, et al. (2012) Contribution of coexisting sulfate and iron reducing bacteria to methylmercury production in freshwater river sediments. Environ Sci Technol 46: 2684-2691.
- Pereira E, Rodrigues S, Otero M, Válega M, Lopes C, et al. (2008) Evaluation of an interlaboratory proficiency-testing exercise for total mercury in environmental samples of soils, sediments and fish tissue. TRAC Trend Anal Chem 27: 959-970.
- Fleck JA, Alpers CN, Marvin-DiPasquale M, Hothem RL, Wright SA, et al. (2011) The effects of sediment and mercury mobilization in the South Yuba River and Humbug Creek Confluence Area, Nevada County, California: Concentrations, speciation, and environmental fate. Part 1: Field characterization, Open-File Report. Reston, VA.
- Gibson B, Ptacek C, Blowes D, Daugherty S (2015) Sediment resuspension under variable geochemical conditions and implications for contaminant release. J Soil Sediment 15: 1644-1656.
- Bloom N, Lasorsa BK (1999) Changes in mercury speciation and the release of methyl mercury as a result of marine sediment dredging activities. Sci Total Environ 237:379-385.
- Eggleton J, Thomas KV (2004) A review of factors affecting the release and bioavailability of contaminants during sediment disturbance events. Environ Int 30: 973-980
- Coelho J, Pato P, Henriques B, Picado A, Lillebø A, et al. (2014) Long-term monitoring of a mercury contaminated estuary (Ria de Aveiro, Portugal): the effect of weather events and management in mercury transport. Hydrol Process 28: 352-360.
- Boetius A, Ferdelman T, Lochte K (2000) Bacterial activity in sediments of the deep Arabian Sea in relation to vertical flux. Deep Sea Res Pt II 47: 2835-2875.
- Smith DC, Azam F (1992) A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar Microb Food Web 6: 107-114.
- Loy A, Lehner A, Lee N, Adamczyk J, Meier H, et al. (2002) Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl Environ Microbiol 68: 5064-5081.
- Lücker S, Steger D, Kjeldsen KU, MacGregor BJ, Wagner M, et al. (2007) Improved 16S rRNA-targeted probe set for analysis of sulfate-reducing bacteria by fluorescence in situ hybridization. J Microbiol Meth 69: 523-528.
- Llobet-Brossa E, Rossello-Mora R, Amann R (1998) microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl Environ Microbiol 64: 2691-2696.
- Pernthaler J, Glöckner FO, Schönhuber W, Amann R 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol 30: 207–226.
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173: 697-703.
- Heuer H, Hartung K, Wieland G, Kramer I, Smalla K (1999) Polynucleotide probes that target a hypervariable region of 16S rRNA genes to identify bacterial isolates corresponding to bands of community fingerprints. Appl Environ Microbiol 65: 1045-1049.
- Heuer H, Wieland G, Schönfeld J, Schönwälder A, Gomes N, et al. (2001) Bacterial profiling using DGGE or TGGE analysis. Horizon Scientific Press, Wymondham, UK.
- Oksanen J, Blanchet F, Kindt R, Legendre P, Minchin P, et al. (2013) Vegan: Community ecology package. R Foundation for Statistical Computing, Vienna.
- Rajapaksha R, Tobor-Kaplon M, Bååth E (2004) Metal toxicity affects fungal and bacterial activities in soil differently. Appl Environ Microbiol 70: 2966-2973.
- Fitzgerald JW (1978) Naturally occurring organosulfur compounds in soil. In Sulfur in the Environment, J Nriagu, Editor. Wiley: New York, pp. 391-443.
- Mora APd, Ortega-Calvo JJ, Cabrera F, Madejón E (2005) Changes in enzyme activities and microbial biomass after “in situ” remediation of a heavy metal-contaminated soil. Appl Soil Ecol 28: 125-137.
- Sinsabaugh RL, Follstad Shah JJ (2012) Ecoenzymatic stoichiometry and ecological theory. Annl Rev Ecol Evol S 43: 313-343.
- King GM, Klug MJ (1980) Sulfhydrolase activity in sediments of Wintergreen lake, Kalamazoo County, Michigan. Appl Environ Microbiol 39: 950-956.
- Burns RG (1982) A D McLaren Memorial Issue. Enzyme activity in soil: Location and a possible role in microbial ecology. Soil Biol Biochem 14: 423-427.
- Lillebø A, Válega M, Otero M, Pardal M, Pereira E, et al. (2010) Daily and inter-tidal variations of Fe, Mn and Hg in the water column of a contaminated salt marsh: Halophytes effect. Estuar Coast Shelf S 88: 91-98.
- Monterroso P, Pato P, Pereira ME, Millward GE, Vale C, et al. (2007) Metal-contaminated sediments in a semi-closed basin: Implications for recovery. Estuar Coast Shelf S 71: 148-158.
- François F, Lombard C, Guigner JM, Soreau P, Brian-Jaisson F, et al. (2012) Isolation and characterization of environmental bacteria capable of extracellular biosorption of mercury. Appl Environ Microbiol 78: 1097-1106.
- Achá D, Pabón CA, Hintelmann H (2012) Mercury methylation and hydrogen sulfide production among unexpected strains isolated from periphyton of two macrophytes of the Amazon. FEMS Microbiol Ecol 80: 637-645.
- Pak KR, Bartha R (1998) Mercury methylation and demethylation in anoxic lake sediments and by strictly anaerobic bacteria. Appl Environ Microbiol 64: 1013-1017.
- Coelho F, Rocha RJM, Pires ACC, Ladeiro B, Castanheira JM, et al. (2013) Development and validation of an experimental life support system for assessing the effects of global climate change and environmental contamination on estuarine and coastal marine benthic communities. Glob Change Biol 19: 2584-2595.
- Porat I, Vishnivetskaya TA, Mosher JJ, Brandt CC, Yang ZK, et al. (2010) Characterization of archaeal community in contaminated and uncontaminated surface stream sediments. Microb Ecol 60: 784-795.
Copyright: © 2015 Oliveira V, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.