Research Article - (2023)Volume 9, Issue 1
Isolation of Cryptococcus laurentii from Mastitic Milk Samples of Cattle and Buffalo in India
Rajesh Chhabra*, G Shrinet, R Yadav, J S Talukdar, N K Kakker and P GoelAbstract
Mycotic mastitis mainly caused by yeasts like Cryptococcus spp. and Candida spp. have been emerging since last decade due to several factors like indiscriminate use of antibiotics, immune suppression, corticosteroid therapy, teat injuries and faulty milking machineries. The fungus, Cryptococcus laurentii is a non-neoformans, encapsulated, basidiomycete, which was earlier considered to be saprophytic and non-pathogenic. Now it is increasingly being reported in humans, especially in the immune compromised patients. In this study, Cryptococcus laurentii was isolated and identified from mastitic milk samples of cattle and buffalo. This probably might be the first report of isolation and identification of Cryptococcus laurentii from mastitic milk samples from India to best of our knowledge. On milk culture examination, typical creamy white color colonies were appeared on Sabouraud's Dextrose agar, which on Gram’s staining gave budding yeast cells appearance. India ink staining revealed bright halo of capsules surrounding the yeast cells. All the isolates were positive forurease production and biofilm formation. Further, confirmation was done using VITEK 2 compact system (BioMerieux) which was based on their biochemical tests profiles. Molecular confirmation was done by the PCR assay. Isolation and identification of this rare fungus from milk samples in present study raises a potential threat of zoonosis.
Keywords
Isolation; Cryptococcus laurentii; Cattle, Buffalo; Mycotic mastitis; Milk sample
Introduction
Bovine mastitis is a multifactorial disease causing huge economic losses to the dairy sector. Mastitis can be caused by over 250 microorganisms which include bacteria, fungi, algae etc [1]. Relative importance of different infections is likely to vary in different geographical regions. In the past, very little importance was given to fungal/mycotic mastitis due to very less percent prevalence but since last decade increasing trend is seen in reports of bovine mycotic mastitis. This could be attributed to several factors like indiscriminate use of antibiotics, immune suppression, corticosteroid therapy, teat injuries and faulty milking machines [2].The most common fungal species isolated from cases of bovine mastitis are the Candida species. However, there have been reports of isolation of different other yeast like Cryptococcus species as well. The genus Cryptococcus is a heterogenous group of encapsulated fungi including important pathogenic species for human and animals, most commonly the Cryptococcus neoformans and non-neoformans Cryptococcus speciesis considered to be nonpathogenic. Eight percent of non-neoformans Cryptococcus species include Cryptococcus laurentii (C. laurentii) and Cryptococcus albidus [3]. Earlier C. laurentii was considered to be saprophytic and non-pathogenic and has also been diagnosed as the etiological pathogen able to cause human infections mainly in immunosuppressed patients [3-5].Many researchers focused on its transmission and effect on immune compromised persons [2-4]. Their findings explored its pathogenicity in multiple hosts and considered it as a pathogen of major concern. C. laurentii has been reported to be a rare and opportunistic yeast pathogen isolated from bovine mastitic milk samples. It has also been associated with biofilm formation and refractory to most of the antibiotic treatments [6]. The present study is probably the first report of C. laurentii isolated from mastitic milk samples of cattle and buffalo from India.
Materials and Methods
Isolation and biochemical identification
The College Central Laboratory (CCL), LUVAS, Hisar, Haryana, India receives milk samples from all over the state of Haryana for routine examination of mastitis. A total of 600 milk samples from buffaloes and cows with a history of chronic mastitis and prolonged antibiotic use were processed for isolation of isolation and identification of rare C. laurentii. Mastitic milk samples were plated onto blood agar, McConkey agar, and Sabouraud dextrose agar and incubated at 37°C for 48-72 hrs. Fungi were identified phenotypically, on the basis of colony morphology and further stained by Gram’s stain and Indian ink capsular stain. The presumptive isolates were also tested for urease activity and biofilm production on Congo red agar plate [7,8].
The isolates were identified and confirmed using VITEK 2 Compact system (bioMerieux) and different biochemical tests results of each isolates were analyzed.
C. laurentii var. laurentii MTCC 2898 (Microbial Type Culture Collection) and Cryptococcus neoformans ATCC 14116 strain (American Type Culture Collection) were used as positive and negative control strains for identification, respectively.
Molecular confirmation of Cryptococcus laurentii
Phenotypically and biochemically confirmed isolates were subjected to DNA samples from all the isolates were extracted using Quick–DNAMiniprep PLUS kit (Zymo Research) and PCR amplification was performed targeting D1-D2 28S rDNA and 18S rRNA gene Internal Transcribed Sequence (ITS) region sequences [9]. PCR using standard procedure was carried out using primer sets as reported earlier in Table 1 [10]. C. laurentii var. laurentii MTCC 2898 and Nuclease Free Water (NFW) were taken as positive control negative control, respectively for PCR assay (Table 1) [11].
| S. No | Target gene | OligoName | 5'<-----Sequence----->3' | Amplicon size(bp) |
|---|---|---|---|---|
| 1 | D1-D2 28S rDNA | CL/28SrDNA/F | GCATATCAATAAGCGGAGGAAAAG | 600 |
| CL/28SrDNA/R | GGTCCGTGTTTCAAGACGG | |||
| 2 | 18S rRNA gene Internal Transcribed Sequence (ITS) | CL/ITS/F | GTCGTAACAAGGTTAACCTGCGG | Variable |
Table 1: Primer sequence for PCR amplification.
Molecular detection of icaA and icaD genes
All the isolates were processed for PCR amplification of biofilm associated genes i.e., icaA and icaD. Standard thermocyler conditions were used for identification of genes. Following primer sets from Table 1 were used for PCR amplification as depicted in Table 2.
| S. No | Target gene | OligoName | 5'<-----Sequence----->3' | Amplicon size (bp) |
|---|---|---|---|---|
| 1 | icaA gene | icaA /F | CCTAACTAACGAAAGGTAG | 1315 |
| icaA /R | AAGATATAGCGATAAGTGC | |||
| 2 | icaD gene | icaD /F | AAACGTAAGAGAGGTGG | 381 |
| icaD /R | GGCAATATGATCAAGATAC |
Table 2: Details of oligo nucleotide primers used for identification of biofilm genes.
Sequence Analysis of Internal Transcribed Sequence (ITS)
The PCR amplified Internal Transcribed Sequence (ITS) products of each isolate were sequenced by Sanger’s Sequencer (Applied Biosystems). The sequences obtained were subjected to nucleotide BLAST (basic local alignment search tool) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to determine the similarity with the already prevalent gene sequences in GenBank (National Center for Biotechnology Information). Twenty nine reference sequences were obtained from public domain of gene bank, NCBI with variable accession numbers. All sequences were aligned using Bio Edit and MEGA6 software to study the variations in the nucleotide sequences and their phylogenetic cluster analysis [12,13].
Results and Discussion
Isolation and biochemical identification
A total of four isolates; three from buffaloes (Isolate number 421, 657, 2085) and one from cow (Isolate number 1848) of C. laurentii were confirmed using phenotypic and biochemical test profile generated by automated VITEK 2 compact system. Automated VITEK 2 compact system revealed that isolate number 421, 657, 1848 and 2085 had 89%, 87%, 90%, and 88% confidence level (probability), respectively of being C. laurentii species. The isolates have typical creamy white colonies on Sabouraud's Dextrose agar (Figure 1a) and blood agar plate. Gram’s staining showed budding yeast cells (Figure 1b) and India ink staining revealed bright halo of capsules surrounding the yeast cells (Figure 1c). All the isolates were detected as biofilm producers (Figure 1d) on Congo red agar plate by producing black colored colonies and also tested positive for urease production(Figure 1e).All of the isolates were found positive for detection of icaA and icaD genes on PCR amplification. Biofilm formation is the important virulence factor of many microorganism including Candida and other fungi which are usually associated with chronic and recurrent infections. There are various methods to detect biofilm production, of them Congo red agar method is most convenient one [14]. Fourteen distinguished biotype patterns were observed on the basis of biochemical test result of each isolates by VITEK 2 compact system from Table 2. Biotype 1 comprised of 26 types of tests which were tested positive by all the isolates. Biotypes 2 to 14 were tested positive by variable numbers of isolates in Table 2.
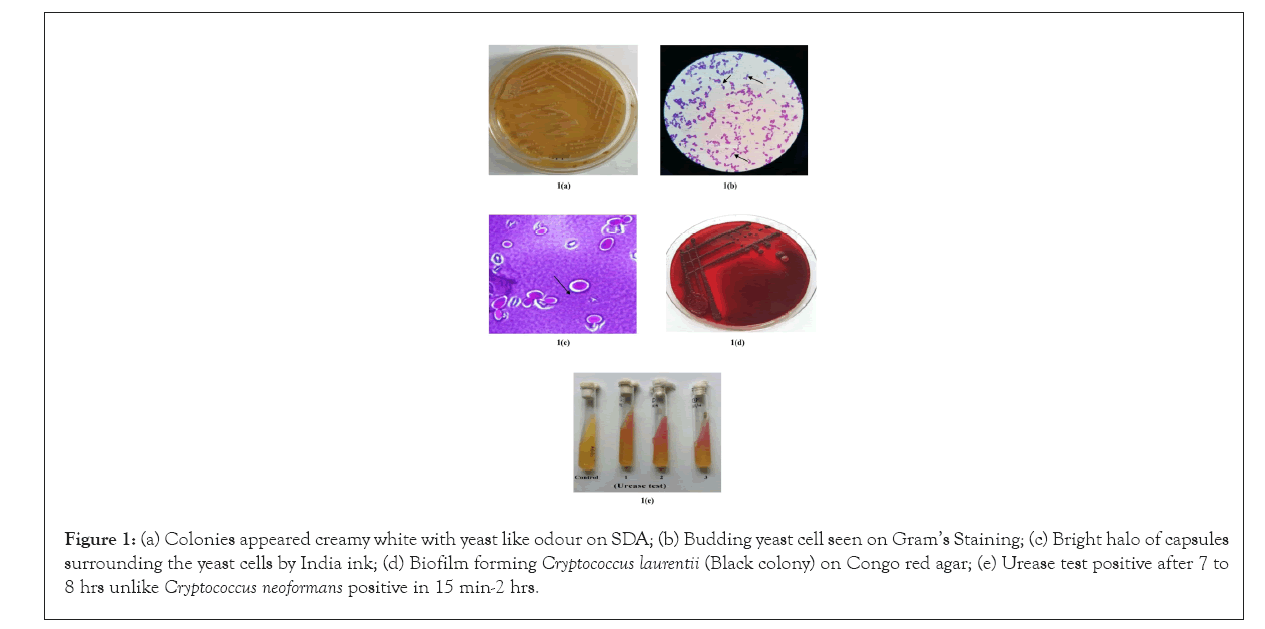
Figure 1: (a) Colonies appeared creamy white with yeast like odour on SDA; (b) Budding yeast cell seen on Gram’s Staining; (c) Bright halo of capsules surrounding the yeast cells by India ink; (d) Biofilm forming Cryptococcus laurentii (Black colony) on Congo red agar; (e) Urease test positive after 7 to 8 hrs unlike Cryptococcus neoformans positive in 15 min-2 hrs.
Molecular confirmation of Cryptococcus laurentii
All the four phenotypically and biochemically confirmed isolates amplified species specific 600bp amplicon size of D1-D2 28S rDNA and 18S rRNA gene Internal Transcribed Sequence (ITS)region sequences producing variable size amplicon of C. laurentii by PCR using primer set reported earlier (Figures 1 and 2, Table 3) [10].
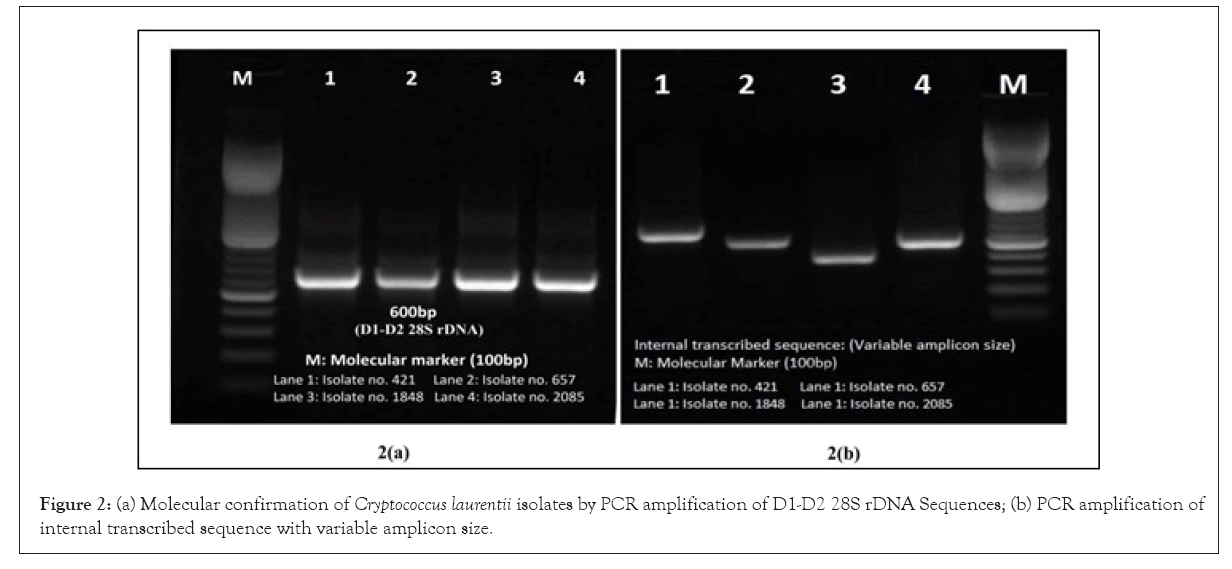
Figure 2: (a) Molecular confirmation of Cryptococcus laurentii isolates by PCR amplification of D1-D2 28S rDNA Sequences; (b) PCR amplification of internal transcribed sequence with variable amplicon size.`
| Biotype | Biochemical Test (VITEK 2) | 421 | 657 | 1848 | 2085 |
|---|---|---|---|---|---|
| 1 | L-Malate assimilation, Leucine-Arylamidase, Arginine, Glycerol assimilation, Arbutin assimilation, Gentobiose assimilation, D-Glucose assimilation, Lactose assimilation, Methyl-a-d-Glucopyranoside assimilation, D-Cellobiose assimilation, D-Maltose assimilation, D-Raffinose assimilation, D-Mannose assimilation, L-Rhamnose assimilation, D-Sorbitol assimilation, Saccharose/sucrose assimilation, D-Turanose assimilation, D-Trehalose assimilation, L-Arabinose assimilation, D-Galacturonate assimilation, D-Xylose assimilation, Acetate assimilation, Citrate (sodium) assimilation, L-Proline assimilation, 2-Keto-D-Gluconate assimilation, D-Gluconate assimilation | + | + | + | + |
| 2 | Alpha-Glucosidase | + | + | + | - |
| 3 | D-Galactose assimilation, D-Melezitose assimilation, Esculin hydrolysis, L-Glutamate assimilation | + | + | - | + |
| 4 | D-Melibiose assimilation | + | + | - | - |
| 5 | Erythritol assimilation, Xylitol assimilation | + | + | + | - |
| 6 | Beta-n-acetyl-glucosaminidase | + | + | - | - |
| 7 | N-Acetyl-Glucosamine assimilation | + | - | + | + |
| 8 | Tyrosine Arylamidase | - | + | + | - |
| 9 | DL-Lactate assimilation, | - | + | - | + |
| 10 | Nitrate assimilation | - | + | - | - |
| 11 | Glucuronate Assimilation | - | - | + | + |
| 12 | L-Sorbose assimilation, Urease | - | - | + | - |
| 13 | Gamma-Glutamyl-Transferase | - | - | - | + |
| 14 | L-Lysine-Arylamidase, Amygdalin assimilation, PNP-N-acetyl-BD-galactosaminidase 1 | - | - | - | - |
Table 3: Biotyping of Cryptococcusl aurentii isolates on the basis of biochemical test results profile generated by VITEK 2 system.
Sequence analysis of Internal Transcribed Sequence (ITS)01
2T2h2e PCR amplified products of 18S rRNA ITS of each isolates were sequenced by Sanger’s Sequencer (Applied Biosystems). All of them showed 100% similarity with ITS sequences of C.laurentii species on nucleotide BLAST at NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Similar sequences with different geographical regions were retrieved from public domain and aligned using Bio Edit [12]. Phylogenetic tree was constructed using MEGA6 by maximum likelihood method based on whole ITS region of C. laurentii and related species with 1000 bootstrap replications [13]. While tracing the sequences for fungi on NCBI, none of them was found to be of Indian origin (Figure 3).
Of the four isolates reported in the present study, two isolates (isolate number 421 and 1848) were in close homology with each other while two isolates (isolate number 647 and 2085) were clustered distinctly in Figure 3. None of them were clustered or grouped with C. laurentii isolates originating from the rest of the world. Phylogenetic analysis revealed their unique identities. All other isolates from USA and European origin were mixed and grouped in separate cluster. To best of our knowledge, none of the isolates were reported earlier from India. Probably this is the first report of C. laurentii isolated from mastitic milk of bovine in India.
Fungal infections, by both yeast and filamentous, are now considered as opportunistic agents causing severe illness in immune compromised hosts (both humans and animals) demonstrating its public health importance. They were previously considered to be non-pathogenic in most of the cases or their pathogenesis was not clearly understood. Various researchers have been working to characterize both most common as well as rare fungal agents in both animals and humans [15,16]. C. laurentii has been associated with fungemia and pulmonary infection in humans [5]. Low birth weight neonate in humans from India has also been reported earlier [2].
Bovine mastitis caused by fungi is considerably less frequent than that caused by bacteria and some-times the infection can become chronic and more difficult to treat. With the wide use of antibiotics across the world, more and more yeast species are being reported from clinical samples. Rare opportunistic fungal species has also been found in milk samples nowadays causing risk to public health [17]. Accurate diagnosis and differentiating bacterial infections from infections in cases of bovine mastitis has become very crucial to minimize potential risks and epidemiological importance. Automated microbial identification such as VITEK 2 compact system has been frequently in use for diagnosis of uncommon fungal pathogens. However their sensitivity and specificity towards variable pathogens may be altered due to numerous reasons. Some organisms are misidentified by commercial automated systems [18]. Modern molecular techniques involving Internal Transcribed Sequence (ITS) region in clinical mycology can yield better accuracy in the etiological analysis of bovine mastitis. The D1/D2 region of 28S rDNA region serves for identification of many genera of yeasts as it is sufficiently variable to recognize species with little nucleotide divergence [10,16,19].
Conclusion
To the best of our knowledge, this might be the first report of rare C. laurentii species from bovine mastitic milk sample in India. The pathogenic potential of the organism is unknown. However, more studies are needed to understand its pathogenicity as primary pathogen causing mastitis in bovines. Additionally, understanding the common patterns of resistance against non-neoformans Cryptococcal infections especially C. laurentii will prevent further treatment failure and economic losses associated with bovine mycotic mastitis.
Acknowledgements
This study was funded by the Lala Lajpat Rai University of Veterinary and Animal Sciences, Hisar.
Conflict of Interest
The authors declare that they have no competing interests.
Funding
“The authors declare that no funds, grants, or other support were received during the preparation of this manuscript”.
Ethics Approval
The milk samples used in the study were directly received in the laboratory from the animal owners for bacterial isolation and antibiotic sensitivity testing. History of the animal with the symptoms of mastitis was recorded at time of sample submission. The milking/milk sample collection procedure does not involve invasive procedure therefore ethical permissions are not indicated. The verbal consent was obtained from the animals owners for the samples under study.
References
- Shome BR, Das Mitra S, Bhuvana M, Krithiga N, Velu D, Shome R, et al. Multiplex PCR assay for species identification of bovine mastitis pathogens. J Appl Microbiol. 2011;111(6):1349-1356.
- Gupta M, Mishra AK, Singh SK. Cryptococcus laurentii fungemia in a low birth weight preterm neonate: India. J Infect Public Heal. 2018;11(6):896-897.
- Khawcharoenporn T, Apisarnthanarak A, Kiratisin P, Mundy LM, Bailey TC. Evaluation of Cryptococcus laurentii meningitis in a patient with HIV infection: A case report and review of the literature. Hawaii Med J. 2006;65(9).
- Cheng MF, Chiou CC, Liu YC, Wang HZ, Hsieh KS. Cryptococcus laurentii fungemia in a premature neonate. J Clin Microbiol. 2001;39(4):1608-1611.
- Banerjee P, Haider M, Trehan V, Mishra B, Thakur A, Dogra V, et al. Cryptococcus laurentii fungemia. Indian J Med Microbiol. 2013;31(1):75.
- Ajesh K, Sreejith K. Cryptococcus laurentii biofilms: Structure, development and antifungal drug resistance. Mycopathologia. 2012;174(5):409-419.
- Türkyılmaz S, Kaynarca S. The slime production by yeasts isolated from subclinical mastitic cows. Acta Vet Brno. 2011;79(4):581-586.
- Silva FA, Medeiros SM, Costa-Junior SD, Roberto AE, Palácio SB, Lima-Neto RG,et al. Antimicrobial resistance profile and biofilm production of microorganisms isolated from oropharynx of rupornis magnirostris (Gmelin, 1788) and Caracara plancus (Miller, 1777). Vet Med Int. 2020.
- Barton R. Laboratory diagnosis of yeast infections.2010:281-309.
- Sugita T, Takashima M, Ikeda R, Nakase T, Shinoda T. Intraspecies diversity of Cryptococcus laurentii as revealed by sequences of internal transcribed spacer regions and 28S rRNA gene and taxonomic position of C. laurentii clinical isolates. J Clin Microbiol. 2000;38(4):1468-1471.
- Vasudevan P, Nair MK, Annamalai T, Venkitanarayanan KS. Phenotypic and genotypic characterization of bovine mastitis isolates of Staphylococcus aureus for biofilm formation. Vet Microbiol. 2003;92(1-2):179-185.
- Hall T, Biosciences I, Carlsbad CJ. BioEdit: An important software for molecular biology. GERF bull biosci. 2011;2(1):60-61.Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725-9.
- Saxena N, Maheshwari D, Dadhich D, Singh S. Evaluation of Congo red agar for detection of biofilm production by various clinical Candida isolates. J Evol Med Dent Sci. 2014;3(59):13234-13239.
- Spanamberg A, Sanches EM, Santurio JM, Ferreiro L. Mycotic mastitis in ruminants caused by yeasts. Cienc Rural. 2009;39:282-290.
- Dalanezi FM, da Paz GS, Joaquim SF, Guimarães FF, Bosco SD, Langoni H. The first report of Cyberlindnera rhodanensis associated with clinical bovine mastitis. J Dairy Sci. 2018;101(1):581-583.
- Krukowski H, Saba L. Bovine mycotic mastitis. Folia Vet. 2003;47:3-7.
- Nath R, Sargiary P, Borkakoty B, Parida P. Cutaneotrichosporon (Trichosporon) debeurmannianum: A rare yeast isolated from blood and urine samples. Mycopathologia. 2018;183(3):585-590.
- Kurtzman CP, Robnett C. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5'end of the large-subunit (26S) ribosomal DNA gene. Journal of clinical microbiology. 1997;35(5):1216-1223.
Author Info
Rajesh Chhabra*, G Shrinet, R Yadav, J S Talukdar, N K Kakker and P GoelCitation: Chhabra R, Shrinet G, Yadav R, Talukdar JS, Kakker NK, Goel P (2023) Isolation of Cryptococcus laurentii from Mastitic Milk Samples of Cattle and Buffalo in India. Appli Microbiol Open Access. 9.242
Received: 19-Dec-2022, Manuscript No. AMOA-22-21002; Editor assigned: 21-Dec-2022, Pre QC No. AMOA-22-21002; Reviewed: 06-Jan-2023, QC No. AMOA-22-21002; Revised: 13-Jan-2023, Manuscript No. AMOA-22-21002; Published: 23-Jan-2023 , DOI: 10.35284/2471-9315.23.9.245
Copyright: © 2023 Chhabra R, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.