
Journal of Clinical and Cellular Immunology
Open Access
ISSN: 2155-9899

ISSN: 2155-9899
Review Article - (2020)Volume 11, Issue 7
Abnormal epigenetic modifications are a trademark of cancer, and endocrine system related organs cancers are without exception. Lately, studies have acknowledged an ever-growing number of epigenetic modifications in both DNA methylation and histone modifications in the endocrine system organs cancers. Novel ultra-deep sequencing and microarray technologies have permitted to study the genome-wide epigenetic patterns in some endocrine system organs cancer types such as parathyroid gland, adrenocortical, and breast carcinomas. However, up till now, in other types of cancer, such as the multiple endocrine neoplasia syndromes, thyroid cancer, the tumor information is scare to candidate genes alone. Future research are required to focus in this direction to cover the deepen understanding of the functional role of these epigenetic modifications in endocrine system cancers, as well as defining their possible clinical utilizations.
Epigenetic modifications; Cancer; Endocrine system; Organs
Adrenocortical carcinoma (ACC) is a rare and often lethal cancer, the frequencies of affecting are about one person per million per year worldwide [176]. About 75% of patients with ACC ultimately develop metastases and improvement on the few accessible standard-of-care medical therapies; outline an incredible need for an improved understanding of the molecular biology of this disease [177]. Even though it has long been known that ACC is characterized by particular histological and genetic features such as high mitotic activity, chromosomal instability, and over expression of IGF2, only in the last two decades of genomics has the molecular landscape of ACC been more thoroughly characterized [178]. The human adrenal cortex, included in adaptive responses to stress, fluid homeostasis, and secondary sexual traits, arises from a tightly regulated development of a zone and cell type-specific secretory pattern [179]. However, the molecular mechanisms controlling adrenal zonation, particularly postnatal zona reticularis development, which produce adrenal androgens in a lifetimespecific manner, remain poorly understood [179]. Epigenetic events, including DNA and histone modifications as well as regulation by noncoding RNAs, are crucial in establishing or maintaining the expression pattern of particular genes and consequently contribute to the stability of a certain differentiation status [180]. Recent evidence points to epigenetic as another regulatory layer that could participate to establishing the adrenal zone-specific pattern of enzyme expression [181]. Ultimately, improved understanding of the epigenetic factors involved in adrenal development and function could lead to novel therapeutic interventions [182].
Histone modifications consider another key mechanism of epigenetic regulation of gene expression [106]. It is known as covalent modifications of histone amino acids. Histone modifications involve enzyme catalyzed reactions at post-translational modifications of N-terminal tails of histone proteins via mechanisms such as methylation, acetylation, phosphorylation and ubiquitinylation. In fact, different types of histone modifications can be occur on a single histone molecule, consequently, increasing combinatorial complexity of post-translational modifications [107].
Gene expression is identified through the pattern of related histone modifications, which is known as the histone codes [108]. Whereas loss of histone acetylation consequences to a denser chromatin conformation with loss of transcriptional activity [109] and histone H3/lysine K4 methylation is associated with active genes. Also, histone methylation can be linked to the methylation of DNA and therefore aberrant expression of gene in cancer cells [110].
The acetylation of histone is found to be regulated via two opposite classes of enzymes. Histone acetyltransferases catalyze the addition of acetyl groups to specific lysine residues of histone and histone deacetylases (HDACs), which removes acetyl groups [111]. The later is the mechanism through which HDACs can control transcription of gene is by regulating acetylation of DNA sequencespecific transcription factors such as E2F, Sp3 and p53. Histone deacetylation has been associated to reduce DNA binding or transcriptional activity. Accordingly, HDACs are considering the vital regulator of cell growth, differentiation and apoptosis [112]. Up to now, there are 18 members of human HDACs enzymes have been documented and classified into four main classes depending on the homology to yeast HDACs in addition to phylogenetic tree analysis [113]. HDACs1–3 are found to be up regulated in ovarian cancer tissues and thought to play a critical role in ovarian carcinogenesis [114].
Recently, a new epigenetic phenomenon has been emerged and defined as posttranscriptional gene down regulation through small non-protein-coding RNA molecules called microRNAs (miRNAs) [115]. MiRNA are known bind to the 3′ UTR region of targeted mRNA resulting in rapid inhibition of the mRNA translation [116] and afterward through the formation of RNA-induced silencing complexes [117]. This biological process causes degradation of the mRNA transcript [118]. In a number of cases, miRNA can also enhance the degradation of the targeted mRNA transcript [119]. This mechanism of genetic ruling is crucial in the underlying processes of cell growth and differentiation. Although miRNAs are vital in normal cell function [120], their aberrant expressions have been associated to the development of cancer [121]. Thus, miRNA expression profiles are promising to be the key expectations for cancer diagnosis and treatment [122].
Newly, miRNAs expression profiles have been documented in ovarian tumors, for example miR-141, miR-200a, miR-200b, miR- 200c and have been found to be up regulated in ovarian cancer tissues [123]. On the other hand, miR-140, miR-145, miR-125b1, and miR-199a, were found to be most miRNAs down regulated among above mentioned miRNAs [122]. Moreover, it was thought that miRNA signatures of ovarian cancer might also distinguish these tumors based on ovarian cancers histologic subtypes: low and high-grade malignancies [124]. Significant down regulation of miR- 143, miR-145 and miR-34c as well as up regulation of miR-29b and miR-29a are found in patients with BRCA1/2 abnormalities when compared to those of lacking demonstrable BRCA1/2 aberrations [125,126]. Furthermore, decreased expression of miR-34b*/c was found to be critical in the progression of ovarian cancer at late stages [126]. Based on the above mentioned information, it became obvious that miRNAs play critical roles in both normal and pathologic ovarian function via targeting the expression of specific genes.
A variety of genes involved in the regulation of cell proliferation and invasion in addition to specific genes in thyroid differentiation are epigenetically modified in thyroid tumor are well acknowledged [127]. For examples of those genes are thyroid transcription factor-1 (TTF-1) [128], TIMP3 [129], RASSF1A [130] and PTEN [131]. Cumulative epigenetic modification play a critical role in the progressions from indolent well-differentiated thyroid carcinomas (WDTC) to metastasizing carcinomas, via a spectrum of poorly differentiated to undifferentiated thyroid carcinoma.
Aberrant DNA methylation of protooncogenes and tumor suppressor genes are reported in thyroid cancer, and it found to occur in other human cancers. Certain specific tumor suppressor genes in the thyroid are DAPK, PTEN, RAPβ2, RAP1GAP, RASSF1A, SLC5A8, and TIMP3.
PTEN was known as a tumor suppressor gene, it is mutated in several types of cancers. PTEN encodes phosphatidylinositol 3, 4, 5 triphosphate 3 phosphatase protein. This gene is negatively regulates signaling pathway of the AKT/PKB and is plays a critical role in the regulation of cell cycle, opposing cell rapid cell division and growth [132]. Aberrant DNA methylation of PTEN is frequently reported in FTC and PTC [131].
The RASSF1A gene encodes a protein similar to the RAS effectors protein (28). The deregulations of RASSF1A mRNA expression are associated with cancer, and aberrant DNA methylation has been acknowledged as a central mechanism underlie in the inactivation of this gene [130,133]. In contradiction of FTC, only a small ratio of PTC harbored the aberrant methylation of RASSF1A, which may play a critical role in thyroid carcinogenesis, independent of the BRAF/MAPK kinase (MEK) MAPK pathway [133].
TIMP3 is a tissue inhibitor of metalloproteinase enzyme, which inhibits the cell growth, invasion, angiogenesis, and metastasis of numerous tumors [134]. TIMP3 gene has been found to be hyper¬methylated in thyroid cancer [129,135]. It is also associated with lymph node metastasis and extra thyroidal invasion [135]. The RAP1GAP gene encodes a type of GTPase activating protein which decreases the activity of the RAS related protein. RAP1GAP is involved in the regulation of oncogenic and mitogenic pathways in thyroid cells [136,137].
RAP1 plays a vital role in the activation of the BRAF MEK ERK pathway and regulation of the ERK dependent pathway [138- 140]. The immunohisto¬chemistry method findings revealed the downregulation of RAP1GAP gene in PTC [141] associated with its proliferation and invasion in thyroid tumor cell lines [142]. In addition, DNA hypomethylation plays a crucial role in carcinogenesis; yet, its role is not clearly understood. Only one study reported the global patterns of aberrant DNA methylation in thyroid cancer subtypes via DNA methylation arrays [143]. 262 and 352 genes were found to be hypermethylated in PTC whereas, 13 and 21 genes were hypomethylated in FTC. Additionally, 280 and 393 hypo¬methylated genes and 86 and 131 hypermethylated genes were determined, which were identified in anaplastic and MTC, correspondingly. Among these genes, four oncogenes including DPPA2, INSL4, NOTCH4 and TCL1B were commonly regulated by hypomethylation [143].
Unluckily, little studies about the histone modifications present in thyroid cancer and the association between those modifications and thyroid toumors behavior is presently available. However, recently, only one study reported global levels of histones acetylation alteration in thyroid cancer tissues [144]. They found that the levels of acetylated H3 at residue K18 were lower in undifferentiated cancers with respect to differentiated cancers, signifying that acetylation is switched off in the thyroid tumor transition. Hypermethylation of the thyroid transcription factor-1 (TTF-1), that is critical for thyroid carcinogenesis, concomitantly with decreased acetyl-H3-K9 and increased dimethyl-H3-K9, has been seen in a subset of thyroid cancer cells that had lost the expression of TTF-1 [128]. Furthermore, it has recently been confirmed that the enhancer of zeste homolog 2 (EZH2), a histone lysine methyltransferase belonging to the polycomb group protein family, is particularly upregulated in ATC, and it directly plays a critical role to transcriptional silencing of PAX8 gene and ATC differentiation [145].
MicroRNAs (miRNAs) are a group of small endogenous noncoding RNAs controlling gene expression in various biological processes, including differentiation, proliferation and apoptosis [146]. The alterations in miRNA expression are thought to be a key regulator of cancer progress and development of thyroid cancer [147]. MicroRNAs are short molecules of 19-23 nucleotide that able to block translation of target mRNA or degradation of mRNA through complementary binding to the 3’- untranslated region (UTR) of mRNAs. Increasing evidence has suggested the contribution of miRNAs in human carcinogenesis [148,149]. The deregulation of miRNA expression is proposed to be an essential regulator of malignancies and progression [150]. As a result of its repression effect, deregulation of certain miRNA may possibly lead to the downregulation of tumor suppressor gene and/or upregulation of oncogenes [151]. Thus, these molecular alterations favor cell differentiation, proliferation and apoptosis. Profiling of MicroRNA in human cancers has revealed signatures associated with cancer diagnosis, prognosis, staging, and response to therapy [152,153]. The roles of miRNAs in the development of various types of cancers, differentiated thyroid cancer and various types of thyroid cancers have been reported [147,154]. These findings strongly suggested critical roles for specific miRNAs in the progression and development of thyroid cancer.
The anomalies in the epigenetic regulation of chromatin structure and function can cause aberrant gene expression and cancer initiation and development [155]. As a result, epigenetic therapies plan to reinstate normal chromatin modification patterns via the inhibition of a variety of mechanism of the epigenetic machinery [156]. DNA methyltransferase and Histone deacetylase inhibitors consider the first acknowledged epigenetic therapies; nevertheless, these agents have pleiotropic property and it remains less clear how they direct to therapeutic responses. More newly, drugs that inhibit histone methyltransferases were developed, maybe representing more specific agents.
The above mentioned findings together reveal that complex epigenetic patterns, including DNA methylation, histone modifications and miRNA abnormalities contribute to thyroid cancer progression and drug resistance. The assessing epigenetic modification profile may provide valuable predictive information for thyroid cancer. Accordingly, reversing epigenetic modifications may nearby itself as a promising treatment modality. While genetic deletions, mutations or allelic losses are irreversible, epigenetic abnormalities are potentially correctable and can be reversed [157,158]. In this scenario, a number of drugs that inhibit DNMT or HDAC action are nowadays in clinical practice or under trial. In preclinical studies, different DNMT inhibitors, for instance azacitidine were found to evoke DNA hypomethylation and reverse chemoresistance of platinum-resistant thyroid cancer cells [159,160] laying the fundamental concept for the clinical assessment of DNMT inhibitors for chemotherapy re-sensitization in thyroid cancer patients [161].
Cancer as a epigenetic genetic arises as a result of fundamental defects in the regulation of cell division [162], and its study therefore has significance not only for public health, but also for our basic understanding of cell biology [163]. In the recent review, a literature review the terms “epigenetic” patterns in thyroid cancer, follicular thyroid carcinoma (FTC), medullary thyroid cancer (MTC), anaplastic thyroid cancer (ATC), DNA methylation in thyroid cancer, miRNA expression in thyroid cancer [164] ‘epigenetic patterns in cancer and the current understanding of epigenetic patterns in thyroid cancer was studied [165]. Recently, cancer genomics is provided as a power full performance into genetic alterations and signaling pathways involved in thyroid cancer [166]. In addition, the current researchers showed the importance of epigenetic modifications and the different types of mechanisms in thyroid cancer [167]. Therefore, the thyroid carcinoma conceders as most frequent endocrine tumor of the endocrine organs [168]. As well as its incidence rate has progressively increased over the last years. More than 95% of thyroid carcinoma is derived from follicular cells in which are considered invasive type of tumor [169]. The molecular pathogenesis of thyroid cancer residue to be clarify, despite of activating the RET, RAS and BRAF oncogenes have been well described [170]. The Increasing the evidence from recent studies demonstrates that acquired epigenetic abnormalities participating with genetic alteration results in modified patterns of gene expression and function [171]. A typically DNA methylation has been established in the CpG regions and microRNAs (miRNAs) expression profile recognized in cancer development [172]. Current studies showed the novel genes that have the type of DNA promoter methylation in papillary, follicular, medullary, and anaplastic thyroid cancer [173]. Anaplastic thyroid cancer (ATC), term referring to undifferentiated subtypes, is considered to be potential and associated with poor prognosis [168]. Traditional treatment including surgery, chemotherapy and radioiodine therapy has been used for thyroid cancer [174]. However, these do not provide any significant reduction of the overall mortality rate [175].
DNA methylation, the most commonly studied epigenetic mechanism, is altered in thyroid cancer. Current technological advances have allowed the identification of novel differentially methylated regions, methylation signatures and potential biomarkers. Though, despite recent improvement in grouping methylation alterations in thyroid cancer [162].
Up till now, about 16 candidate tumor suppressor genes have been documented in ovarian cancer. Only 3 of those genes are confirmed to be imprinted, which include: pleiomorphic adenoma genelike 1 (PLAGL1), ARHI (or DIRAS3), and paternally expressed 3 (PEG3) [101]. Those 3 genes are found to be down regulated in a significant fraction of ovarian cancers [102]. DNA hypermethylation of the CpG is one of the projected mechanisms underlying the observed downregulation [103]. In ovarian cancers, downregulation of functional alleles due to promoter hypermethylation has been reported in 10% and 26% of cases for PEG3 [104] and ARHI [102] correspondingly. Treatment of ovarian cancer with histone deacetylase inhibitors and demethylating drugs has been capable to aggravate expression of ARHI and PEG3 genes in ovarian cancer cell lines, which found to be associate with cell growth inhibition [105]. Taken together, these studies propose that cancers treatment with histone deacetylase inhibitors and demethylating agents may provide particularly useful if epigenetic transformations have impacted the imprinted genes. Up regulation of imprinted genes has also been implied in ovarian cancer for instance, over expression of IGF2 is often happening in serous ovarian cancer and the aberrant methylation of the IGF2-H19 locus is proposed as a fundamental event in ovarian tumorigenesis.
Pituitary cancer is a rare type of malignant intracranial neoplasm defined as distant metastasis of pituitary adenoma [183]. Although, the frequency of pituitary adenoma is low because only 0.1% to 0.2% of PAs finally develop into PCs, the diagnosis is poor and 66% of patients die within the first year [184]. Wide majority of pituitary cancer are benign and behave consequently; however, a part are invading and are more aggressive, with a very small part being frankly malignant [185]. The cellular pathways that drive transformation in pituitary neoplasms are poorly described, and recent classification methods are not reliable correlates of clinical behavior [186]. Recently, the techniques in epigenetics research, that investigate the modifications in gene expression without changes to the genetic code [187], provide a new aspect to describe tumors, and may hold implications for prognostication and management [188]. The genetic pattern of pituitary adenomas (PAs) is various and numerous of the identified cases remain not understand pathogenetic mechanism [189].
Recently, the genetic researches have advanced our understanding of pancreatic cancer at a mechanism and translational level [190]. Genetic concepts and tools are increasingly starting to be applied to clinical practice, specifically for accuracy diagnostics and therapy [191]. However, an epigenomics technique is rapidly emerging as a promising conceptual and methodological pattern for advancing the knowledge of this disease [192]. Moreover, the current researches have uncovered potentially actionable pathways, which provide for the prediction that future characterization for pancreatic cancer will involve the potent testing of epigenomic therapeutics [193]. Hence, epigenomics promises to generate a significant amount of new knowledge of both biological and medical importance [194].
Worldwide, lung cancer is the majority cause of cancer associated with mortality [195]. Tumorigenesis includes a many steps process resulting from the interactions of genetic, epigenetic, and environmental factors [196]. Genome-wide association studies and sequencing studies have recognized several epigenetic modifications correlating with the progress of lung cancer [163]. Epigenetic mechanisms, fundamentally including aberrant DNA methylation, histone modification, and noncoding RNAs (ncRNAs), are genetic and reversible modifications that are involved in some essential biological processes and influence cancer biomarks [197]. Recently, the scientific advancements have a permitted to detecting the regions that undergo variety patterns of methylation [198]. It has been elucidated that the dominated on changes in gene expression is not directed only by transcription factors and that epigenetic alterations are also involved in this process [199]. Moreover, epigenetic modifications have been demonstrated to be significantly in cancer progress [200].
In spite of enormous social and scientific attempts, fatness rates continue to promote worldwide [201]. However, genetic factors participate to obesity progress; genetics alone cannot explain the current epidemic [202]. Obesity is fundamentally the consequence of complex genetic-environmental interactions [203]. Proof suggests that contemporary lifestyles trigger epigenetic changes, which can dysregulate energy balance and thus contribute to obesity [204]. The hypothalamus plays a crucial role in the regulation of body weight, via developed network of neuronal systems [205]. Modifications in the activity of these neuronal pathways have been involved in the pathophysiology of obesity [206].
Cancer epigenetic provider to the scholars a visualization both above and beyond the view of genotype (For instance DNA sequencing) influencing the phenotype without intervention of any other key players. Thus, allowing researchers to illustrate the involvedness of interactions accessible from the genes to the cellular fate. The observable fact that the mechanisms can be reversed offers the process a range of flexibility not expected before. This interesting flexibility of biological distinctiveness is also very attractive for both clinical cancer monitoring and management aspects and prognosis as well.
Although a number of genes that undergo aberrant epigenetic deactivation associated with endocrine system organs cancers are growing, a number of questions necessitate to be addressed before we fully understand the biologic implication and outcome of this process. For example, what is the mechanism underlying selective methylation of genes involved in endocrine system organs cancers which increases the activity and expression of DNMTs enzymes? Why do hypomethylation and hypermethylation are take place in in endocrine system organs cancer cells? Is there an active demethylation mechanism to elucidate for hypomethylation? or it is caused by reduced hypermethylation?
Abundance of evidence indicates that the detection of DNA methylation can serve up as a talented cancer biomarker. However, up till now, in endocrine system organs cancers some gene reveals adequate specificity and sensitivity in the detection of in endocrine system organs cancers to assurance further testing as a cancer biomarker in patients’ body fluids. Further researches are strongly recommended to center on identification of novel aberrantly methylated genes by high-throughput screening techniques such as CpG island microarray assays as the gateway in the direction of cancer biomarker confirmation. Moreover, in in endocrine system organs cancers methylation fingerprint comprise several genes may be more precise than that of individual genes in the early diagnosis and risk measurement of in endocrine system organs cancers and molecular detection of reseated specimens.
Prior to completely identify the etiology and consequences of global hypomethylation in in endocrine system organs cancers, the therapeutics targeting DNMTs in cancer must be used with caution. Ideal therapeutics is that can selectively activate a set of methylated genes without unwanted demethylating to the rest of the genome. Known the close association between histone deacetylation and DNA methylation in epigenetic deactivation, a combination of both DNMT inhibitors and histone deacetylation inhibitors may be an successful strategy for the treatment of in endocrine system organs cancer patients.
The authors declare that there is no conflict of interests.
The authors thank Mr. Yagoub Musa Adam for provide us the free internet.
DNA methylation refers to the addition or removal of a methyl group to a cytosine residue in the DNA sequence [1]. DNA methylation is controlled by DNA methyltransferase enzymes [2]. In genome wide association studies, overall reduction of DNA methylation (what so called DNA hypomethylation) are the main functionally associated when they occur in transcriptional regions of genes [3], resulting in different levels of transcription [4]. It is currently known that DNA hypomethylation plays a fundamental role in carcinogenesis via favoring mitotic recombination [5], leading to translocations, deletions, and chromosomal remodeling. The addition methyl groups (What so called DNA hypermethylation) is much more gene specific [6]. The sequences of DNA that are enriched for cytosine preceding a guanine (CpG dinucleotide) sites are called CpG islands which mainly located in the promoter region [7]. In particular, CpG islands occur in promoters of about half of all genes [8]. Hypermethylation of promoter CpG islands, that are approximately contains all active genes may lead to transcriptional silencing and consequently lead to a reduction in protein expression [9] (Figure 1). Therefore, DNA hypermethylation of tumor suppressor genes is now documented as a process of silencing alternative to mutation or allelic loss in cancer development [10].
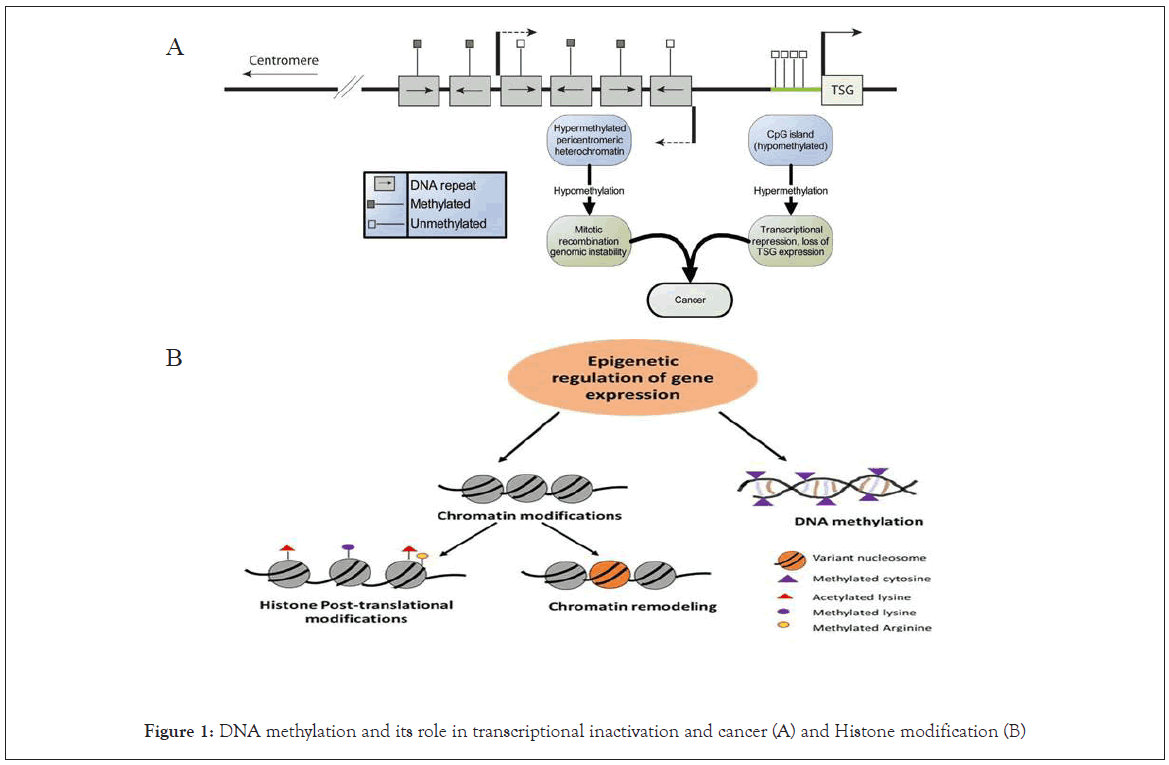
Figure 1: DNA methylation and its role in transcriptional inactivation and cancer (A) and Histone modification (B)
It is well documented that the hypermethylation of genes includes DNA repair, cell, carcinogen metabolism, program cell death, and cell-cell interactions [11], which have been implicated in carcinogenesis [12]. Hypermethylation can also reduce the transcription of microRNAs leading to induction of carcinogenesis [13]. However, it should be distinguished that DNA hypermethylation also occurs in normal physiological process [14], for example, during inactivation of the second X chromosome in females. The question is raised up is why aberrant DNA methylation occurrences is not fully understood. It was found that the DNA methylation profile of a number of genes is age dependent manner [15,16], while others are genes are methylated in a cancer-specific manner [17]. A carcinogenetic pathway of particular gene that is relevance to the reproductive system is the CpG island methylator phenotype (CIMP) dependent. CIMP+ cancers have different genetic, pathologic and clinical profile. Whether environmental factors such as carcinogens, diet (e.g., folic acid), or other unidentified causative agents contribute to DNA methylation still to be clarified and it's an area of interest research to be conducted.
In 1999, two dissimilar types of colorectal cancers were documented and found to demonstrate low and high levels of tumor-specific methylation correspondingly. The latter type of cancer was found to display a “CpG island methylator phenotype” (CIMP) [89]. CIMP was reported to be negatively linked with genetic abnormalities in colorectal cancer [90,91] which imply that it can offer an alternative biological pathway for carcinogenesis [92]. Other cancers types which illustrate frequent and attendant deactivation of a number of genes via hypermethylation have also been designated as CIMP. Those cancers involve liver, gastric [93], leukaemia [94], lung, and ovarian cancers [95]. Differential expression of DNA methyltransferase (DNMT) genes in some ovarian cancer cell lines has been observed [96] and the modifications in DNMT mRNA expression might contribute to the development of the CIMP phenotype in ovarian cancer. However, the central mechanisms that contribute to aberrant DNA methylation in ovarian cancer cells are still less clear.
Several studies established that patients with CIMP positive cancers have poorer prognosis probably due to the increases of their epigenetic plasticity. Therefore, it is of importance to establish the molecular basis of CIMP and confirm the present set of methylation biomarkers for CIMP detection. However, CIMP positive tumors are easier to be diagnosed at an early stage due to the aberrant DNA methylation which can be detected with high sensitivity [97]. Also, this phenotype may be competent to forecast outcome of medication in ovarian cancer patients [98,99]. In general, the compactness of methylated CpG sites within a locus in addition to the number of methylated loci were found to increase in late stages of ovarian cancer [100]. Although the duration of progression-free survival following chemotherapy treatment was found to be significantly shorter for patients with high levels of DNA methylation compared to those with lower DNA methylation levels detection of CIMP tumors which may aid in treatment planning and outcome.
Ovarian cancer comprises the most serious gynecological disease and the 7th top cause of cancer death in female [83]. Unluckily, patients with ovarian cancer are routinely diagnosed at late stage of cancer progression, when the cancer has been already spread into the abdominal cavity and complete surgical removal is not achievable. The prediction of an individual who at a higher risk of developing ovarian cancer possibly may permit lifestyle interventions [84], chemotherapy prevention [85,86]. The etiology of ovarian tumor is not well defined. Yet, general modulators of ovarian cancer risk include increasing of age [87] and reproductive function history [88]. Earlier pregnancies, increased number of pregnancies, oral contraceptives usages, tubal ligation in addition to hysterectomy have all been confirmed to be associated with a lower risk of ovarian cancer. To determine the epigenetic factors fundamental in this protective effect, it is of importance to have a accurate detection of the normal progenitor cells from which ovarian cancer develops. Nevertheless, the compromise on the initial point of ovarian cancer has not yet been approved. Normal human ovarian epithelial cell lines have been extensively used in laboratory research approach. Lately, the fallopian tube cells implanted in the ovary or inclusion cysts have been projected as progenitors of ovarian cancerous cells. Additionally, a mullerian basis for ovarian cancer cells has also been argued for.
The miRNAs are small, noncoding molecules of RNAs consist of 18-22 nucleotides, bind to the 3' untranslated region of mRNAs [62]. They play a critical roles in post-transcriptional degradation or inhibition of target mRNA, depending on the level of complementary base pairing [63-65]. Thus, miRNAs play a vital role in the regulation of many cell and tissue functions for instance, cell differentiation, development, metabolism and cell cycle [66- 68]. Aberrant expression of miRNAs is found to have critical impact on a variety of biological processes such as infection and cancer development [66,69,70].
The role of miRNAs in cellular differentiation, growth, and apoptosis of cancer cells via their targeting mRNA has been reported [71-73]. MiRNAs may be tumor suppressors or oncogenic molecule [74,75], with tumor suppressors being downregulated whereas, oncogenic being upregulated in cancers. Commonly, the roles of miRNAs in the cancer has been emphasized by the fact that approximately 50% of all miRNA genes are located in the so called 'fragile sites', the cancer associated genomic regions that are frequently altered in cancer. A bulk of data has already been documented about aberrant expression of miRNAs in the cancers. The understanding of the functional consequences of these abnormalities have not been molecularly developed [76].
The role of miRNAs in prostate cancer is getting clearer via understanding the interactions between miRNAs and their targeted mRNAs and the consequential impact on carcinogenesis of the prostate cancer [77,78]. It is found that miRNAs and their targeted mRNA are aberrantly expressed in prostate cancers which, in turn, change the cellular invasion, growth, and metastatic potential of prostate cancer cells. The deregulations of certain miRNAs expressions are now considered crucial biomarkers for classification, prognosis and diagnosis, of prostate cancer [75,79,80]. All above mentioned data highlights the significance of the biology of miRNAs in prostate cancer. Their specific deregulations, and how one could control their expressions will possibly become novel roads through which newer therapeutic strategies might be developed for the medication of metastatic castrate resistant prostate cancer. Several types of miRNA play a crucial role in prostate cancer, for example, miRNA-488 [81], miRNA -221 and miRNA-222 [82].
Histone modifications play a critical role in normal and cancer cells, thereby orchestrating key of physiological and pathological processes [60]. In prostate cell lines, methylation of lysine 9 in histone 3 (H3K9) is associated with histone H3K4 methylation and repression of androgen receptor (AR) genes [47] is linked with AR gene activation in castration-resistant prostate cancer (CRPC) cell lines and tissues [47]. H3K4 is considerably methylated at the androgen receptor enhancer of the protooncogene (Ubiquitinconjugating enzyme E2C) UBE2C gene in CRPC, which cause androgen receptor binding and expression of UBE2C mRNA [47]. Heat shock protein 90 is also found to play a critical role in androgen-induced and -independent nuclear localization and activation of AR. Histone deacetylase 6 (HDAC6) regulates AR hypersensitivity and nuclear localization, mainly through modulating Tumor Necrosis Factor Receptor-Associated Protein-1 (TRAP1) acetylation [61].
The majority of studies focused on DNA hypermethylation have been done in epigenetic field of prostate cancer [52]. Certainly, gene-silencing is found to be more frequent by hypermethylation of promoter region compared to mutations in carcinogenesis [53]. Many studies on different hypermethylated genes in different types of cancers propose the main part of the carcinogenesis [54].
At present, more than 100 genes have been detected for their frequencies of hypermethylation and for their critical role in prostate carcinogenesis [55-59]. Several of those hypermethylated genes are tumor suppressor genes that code for the proteins that control the cell cycle and/or endorse apoptosis. The roles of tumor suppressor genes in prostate cancer undergo into five main categories: cell cycle, DNA repair, apoptosis, invasion/metastasis, and corticosteroid hormonal response. Loss function of these genes via promoter hypermethylation ultimately contributes to the carcinogenesis and pathogenesis of prostate cancer.
Hypomethylation is the most methylation problem that is seen in a wide variety of cancers including prostate cancer [49]. Hypermethylation alterations seem to precede hypomethylation alterations, which are usually detected in cancers of late, stage and occur heterogeneously during prostate cancer development and metastatic dissemination [47]. Hypomethylation has been proposed to contribute to oncogenes is via several mechanisms including: activation of oncogenes for instance H-RAS and c-MYC, activation of latent retrotransposons, and through contributing to chromosome unsteadiness [50]. Modern reports have confirmed strong association between MYC upregulation in prostate cancer cell and clinical progression [51]. MYC plays a crucial role in androgen-dependent growth and subsequent its ectopic expression can stimulate androgen-independent growth in prostate cancer tissue [47].
Prostate cancer is one of the most frequent human malignancies and driven via genetic and epigenetic modifications [48]. Epigenetic modifications include DNA methylation, histone modifications, and microRNAs (miRNA) which produce heritable alterations in gene expression without changing the DNA sequence. Aberrant DNA methylation (hypomethylation and hypermethylation) is the most excellent characterized alteration in prostate cancer and leads to instability of genome and improper gene expression. Global and locus-specific alterations in chromatin rearrangement are associated with prostate cancer, suggesting a causative malfunction of histone-modifying enzymes. In addition, microRNA deregulation contributes to the carcinogenesis of prostate cancer, including interference with apoptosis and androgen receptor signaling pathways. There are significant relations between common genetic alterations and the epigenetic landscape change.
Prostate cancer is one of the most widespread cancers affect men with >1,100,000 new cases and 300,000 annual deaths globally [45]. The disease is more frequent among overage men, with a middle age at diagnosis about age above 60 years [45]. Prostate cancer is the main medical problem that requires urgent attention as the disease is indolent, shows prolonged latency in association with high morbidity and mortality [46]. Epigenetic modifications have found to plays critical roles in prostate cancer development and metastasis [47].
Genomic imprinting is one of epigenetic mechanism inducing functional differences between maternal and paternal genomes that play crucial role in mammalian embryonic development. The differential methylation of CpG islands in critical regions of imprinted genes is the mechanism of the imprinting progression that differentiates maternal and paternal alleles [37]. Maternally expressed H19 is one of the most excellent-characterized imprinted genes. Expressions of imprinted genes, for instance H19, in the germ line become biallelic at day 11.5 during the embryonic development [38,39].
The 5′-region of H19 is usually methylated on the paternal allele in human genomes [40]. It has been shown that fetal spermatogonia are mainly unmethylated at the differentially methylated CpG regions of H19, while adult testicular germ cells show significant methylation at the same CpG regions [41]. These phenomena are considered as “DNA reprogramming”, which are seen in genomewide of germ cells or pre-implantation embryos [42]. It has been also found that the complete epigenetic reprogramming occurs within 1 day of the embryonic developmental period in mouse primordial germ cells after reaching the genital crest [41,43]. In addition, loss of allele-specific imprinted gene methylation has also been seen in both male and female primordial germ cells of the mouse [44]. These data support the notion that pre-existing imprints are erased in the germ line by this stage.
The most important role of histone acetylation during the spermatogenesis is the histone H4 hyperacetylation during spermiogenesis [34]. This process seems to play a critical role for subtraction of histones and their replacement via protamines, which is a fundamental characteristic for nucleus condensation, and consequently formation of spermatozoa. There are several members divided in different subfamilies. Interestingly, choriocarcinomas displayed commonly high expression for all three classes I of HDAC isoforms [35]. In contrast with other types of cancers, no diagnostic or predictive values for HDAC1–3 in testicular cancer could be inferred [36].
The major role of epigenetic modifications has been reported in carcinogenesis. Certainly, it has been found that DNA methylation is linked with repression the expression of tumor suppressor gene. This epigenetic alteration is one of the most frequently studied in the cancer research field, and has been documented as a major mechanism during testicular cancer progression [28,29]. The DNA methylation patterns seem to be associated with histological characteristics of the different types of testicular cancer. Dnmt1 was not expressed in seminoma, however it over expressed in embryonic carcinoma [30]. In contrast, the expression of Dnmt3a was found to be over expressed in testicular cancer compared to non-tumor testicular tissues [31]. The pattern of Dnmt3b expression has been extensively studied and demonstrated that it could be used as a prognostic biomarker for relapse of stage I seminomas [32] In addition, Dnmt3l was upregulated in the non-seminoma tumors [33].
DNA methylation plays a critical role during germ cell development. These epigenetic modifications rely on DNA methyltransferases (DNMTs). Among these enzymes, the expression profiles of DNMT3a and DNMT3l propose that they may function during embryonic germ cell development for the organization of de novo DNA methylation. On the other hand, DNMT1 and DNMT3b are found to increase apparently following the birth in male [27]. Thus it is assumed that these two enzymes may be implicated in the maintenance of DNA methylation patterns in the proliferating of spermatogonia.
Testicular tumor (TC) is unusual cancer which comprises 1% of cancers in men. Nevertheless, it is the most frequent tumor in men in their 20s and 30s [24]. It is a real sterile problem which affects men throughout their reproductive period. Interestingly, the development of testicular cancer is associated with urogenital disorders [25]. Testicular cancer development also has been linked to hypospadia, cryptorchidism and low fertility. In fact, epidemiological studies dispute for an increased risk of testicular germ cell tumor in males who experience with fertility troubles [26].
In the last 20 years, advances in the field of endocrine oncology have enabled the genetic basis of some hereditary endocrine tumors to be uncovered, and they have also contributed to increasing knowledge of certain sporadic diseases, and consequently the development of new diagnosis or treatment methods. In addition, the contribution of epigenetic mechanisms in tumor development has been widely described. In this review, we focus on those endocrine tumors where the role of certain epigenetic mechanisms (DNA methylation and histone modifications) has been demonstrated. Endocrine tumors affect parts of the body that secrete hormones and include adrenal gland tumor (adrenocortical carcinoma, ACC), islet cell tumors (gastrinoma, VIPoma, glucagonoma, and somatostatinoma), neuroendocrine tumors (such as pheochromocytoma), as parathyroid and thyroid carcinomas, among others. In the following sections, for the purpose of discussion, these endocrine tumors will be divided into those which are hereditary (multiple endocrine neoplasia (MEN) syndromes) and those which are sporadic (thyroid, parathyroid, breast and ovarian and prostate adrenocortical and lung neuroendocrine tumors and pheochromocytoma, and paraganglioma).
Histones are the protein biomolecules of chromatin, that the DNA is wound around (Figure 1). Histones can subjected to a number of diverse of post-translational modifications. These modifications include methylation, acetylation [18], phosphorylation [19], and ubiquitination [20]. Significantly, these post-translational modifications can affect the interactions between histones and DNA, resultant in changes of gene transcription, DNA replication, chromosomal organization and DNA repair [21]. Generally, Acetylation of histones is associated with activation of transcriptional [22]. Therefore, the deacetylation is involved in tumor suppressor genes silencing, which is critical in carcinogenesis [23]. Certainly, histone deacetylase enzyme inhibitors are fundamental in early phase clinical trials for the medication of various types of cancers, which show potential results and promising of future cancer treatment.
Citation: Ahmed AA, Essa MEA, Mohammed FA, Mohamed ASS (2020) Epigenetic Modifications in the Endocrine System Organs Cancer. J Clin Cell Immunol. 11:605.
Received: 29-Sep-2020 Accepted: 13-Oct-2020 Published: 20-Oct-2020 , DOI: 10.35248/2155-9899.20.11.605
Copyright: © 2020 Ahmed AA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.