Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research - (2024)Volume 17, Issue 1
Background: The PKHD-1 (Polycystic Kidney and Hepatic Disease-1) gene encodes a crucial protein vital for renal and hepatic functions. Mutations in PKHD-1 result in Autosomal Recessive Polycystic Kidney Disease (ARPKD), a severe disorder in early infancy. Despite its significance, the structural information on PKHD-1 remains limited, with few low-resolution structures accessible. Homology Modeling was employed to generate structural models of PKHD-1 proteins from three species: Homo sapiens (Human), Mus musculus (Mouse) and Canis lupus familiaris (Dog). Various bioinformatics tools were utilized for analysis and validation.
Results: Structural models of PKHD-1 proteins from different species were generated using Homology Modeling and advanced bioinformatics tools, including SWISS-Model, ProtParam, GOR4, Protein Structure Analysis (PROSA) Web, ExPasy QMEANDisCo and P2Rank. The primary structure, physicochemical properties and secondary structure of PKHD-1 proteins were analyzed and validated. Binding pockets critical for understanding functional roles and potential therapeutic interventions were predicted using the P2Rank tool.
Conclusion: This study provides comprehensive structural insights into PKHD-1 proteins across multiple species. Rigorous validation of homology models through Z-Score analysis and QMEANDisCo Global Score ensures their reliability and accuracy. The identification of binding pockets offers potential targets for therapeutic interventions. Comparative analysis of PKHD-1 protein structures enhances understanding of evolutionary relationships and lays the foundation for future comparative functional studies. This research significantly contributes to structural biology and biomedical research, serving as a valuable resource for researchers investigating PKHD-1 function, disease mechanisms and drug targeting strategies. The findings pave the way for exploring species-specific functions and adaptations of PKHD-1, fostering advancements in the understanding and treatment of ARPKD and related disorders.
PKHD-1 protein; Autosomal Recessive Polycystic Kidney Disease (ARPKD); In silico analysis; Bioinformatic tools; Homology modeling
The structural elucidation of proteins is a fundamental endeavor in molecular biology, providing crucial insights into their functions, interactions and potential therapeutic targets. Protein structure determination has been a cornerstone of research in the life sciences, offering profound contributions to our understanding of biological processes. The protein PKHD-1 stands as a prominent yet enigmatic member of the protein world. The primary sequence of PKHD-1 has been well-documented; however, its three-dimensional structure remains elusive.
The PKHD-1 protein plays a pivotal role in renal and hepatic development and its dysfunction has been associated with the pathogenesis of ARPKD in humans, a severe condition with limited treatment options [1]. PKHD-1, a large and complex protein, is primarily expressed in renal and hepatic tissues, contributing to the development and maintenance of these vital organs [2]. Furthermore, recent studies have emphasized the importance of PKHD-1 in liver function and biliary homeostasis in mice [3].
The comparative analysis of PKHD-1 among different species, such as mouse, human and dog hold significant promise for unraveling its structure-function relationships and evolutionary conservation. Homology modeling, a computational technique for predicting protein structures based on the alignment of target protein sequences with known structures, offers a potent approach to bridge this gap [4].
Moreover, the shared and distinct functions of PKHD-1 in these species are of immense interest in the context of comparative biology. While mouse models have been invaluable for studying the genetic basis of PKHD-1related disorders and the associated developmental abnormalities, human studies have revealed the clinical implications of PKHD-1 mutations in ARPKD [3,5]. Additionally, the role of PKHD-1 in the pathogenesis of hepatic disease in dogs has not gone unnoticed [6].
In silico predictive homology modeling has proven to be a powerful tool for deciphering protein structures when experimental methods are challenging, costly or unfeasible. It leverages the homologous regions of well-characterized proteins to generate 3D structural models for the protein of interest. To date, the structure of PKHD-1 has not been successfully determined experimentally, underscoring the necessity for computational approaches like homology modeling.
A comprehensive investigation into PKHD-1 across these three species-mouse, human and dog-can elucidate both commonalities and species-specific variations in its structure and function. Such insights may hold the key to understanding its diverse roles in renal and hepatic biology and inform the development of novel therapeutic strategies.
PKHD-1 protein sequences of mouse, human and dog
The PKHD-1 protein sequences of M. musculus, H. sapiens, C. lupus familiaris were retrieved from UniProt, a comprehensive resource for protein sequence and annotation data (Table 1) [7]. The Accession No. was E9PZ36 for M. musculus, P08F94 for H. sapiens and E2RK30 for C. lupus familiaris.
| Protein name | Length of sequence | UniProt ID | Organism |
|---|---|---|---|
| PKHD1 | 4059 | E9PZ36 | Mus musculus |
| PKHD1 | 4074 | P08F94 | Homo sapiens |
| PKHD1 | 4074 | E2RK30 | Canis lupus familiaris |
Table 1: Protein sequence retrieved from UniProt.
Physico-chemical characteristics
The physical and chemical characteristics of protein [molecular weight, theoretical pI, amino acid composition, atomic composition, formula, extinction coefficients, estimated half-life, instability index, aliphatic index and Grand Average of Hydropathy (GRAVY)] of PKHD-1 proteins were computed by ProtParam tool (Tables 2-3) [8].
| S.no | Name of organism | Mol. wt. | pI | EC (assuming all pairs of Cys residues form cystines) | EC (assuming all Cys residues are reduced) | Half-life (hrs) |
|---|---|---|---|---|---|---|
| 1 | M. musculus | 444882.1 | 5.9 | 515480 | 509480 | 30 |
| 2 | H. sapiens | 446701.72 | 6.12 | 502780 | 496530 | 30 |
| 3 | C. lupus familiaris | 447576.09 | 5.95 | 529460 | 523460 | 30 |
| S.no | Formula | II | GRAVY | -R | +R | AI |
| 1 | C19871H31069N5349O5930S159 | 41.35 | -0.012 | 379 | 312 | 91.62 |
| 2 | C19902H31204N5430O5919S170 | 44.73 | -0.02 | 367 | 313 | 92.43 |
| 3 | C20005H31327N5393O5937S162 | 45.24 | -0.003 | 373 | 312 | 93.9 |
Note: Mol. wt.: Molecular weight; pI: Isoelectric point; -R: Number of negative residues; +R: Number of positive residues; EC: Extinction coefficient at 280 nm; II: Instability index; AI: Aliphatic index; GRAVY: Grand Average of Hydropathy.
Table 2: Physicochemical properties of PKHD-1 protein.
| S.no | Amino acids | M. musculus | H. sapiens | C. lupus familiaris |
|---|---|---|---|---|
| 1 | Ala (A) | 5.90% | 5.60% | 5.60% |
| 2 | Cys (C) | 2.40% | 2.50% | 2.40% |
| 3 | Asp (D) | 4.20% | 3.90% | 4.10% |
| 4 | Glu (E) | 5.20% | 5.10% | 5.10% |
| 5 | Phe (F) | 4.40% | 4.20% | 4.20% |
| 6 | Gly (G) | 7.60% | 8.00% | 7.7& |
| 7 | His (H) | 2.60% | 2.80% | 2.50% |
| 8 | Ile (I) | 5.20% | 5.80% | 6.70% |
| 9 | Lys (K) | 3.30% | 3.10% | 3.20% |
| 10 | Leu (L) | 10.40% | 10.00% | 10.00% |
| 11 | Met (M) | 1.60% | 1.70% | 1.60% |
| 12 | Asn (N) | 4.20% | 4.80% | 4.60% |
| 13 | Pro (P) | 5.40% | 5.30% | 5.50% |
| 14 | Gln (Q) | 4.20% | 4.50% | 4.50% |
| 15 | Arg (R) | 4.40% | 4.60% | 4.50% |
| 16 | Ser (S) | 9.60% | 9.20% | 9.50% |
| 17 | Thr (T) | 6.80% | 6.40% | 6.20% |
| 18 | Val (V) | 8.60% | 8.60% | 8.10% |
| 19 | Trp (W) | 1.60% | 1.60% | 1.60% |
| 20 | Tyr (Y) | 2.50% | 2.40% | 2.60% |
Table 3: Amino acid composition.
Secondary structure predictions of PKHD-1 protein
The secondary structure predictions of the PKHD-1 protein were made by employing GOR4 [9].
PKHD-1 protein model building and evaluation
The linear amino acid sequence of PKHD-1 protein of mouse, human and dog were retrieved from UniProt protein sequence database [7]. The template search for tertiary structure was performed against SWISS-MODEL Template Library [10]. After optimization the 3D model were verified using the MolProbity and PROSA programs [11]. PROSA web server is a web-based tool applied for the validation of the modeled protein structure with available protein structures from Protein Data Bank (PDB) on the basis of Z-Score [12]. MolProbity server is used for validation of all-atom structure and plotting Ramachandran plot [13].
Binding pocket prediction
The binding pockets of PKHD-1 protein in all three species (M. musculus, H. sapiens, C. lupus familiaris) were predicted using P2Rank tool (PrankWeb web server) [14-16].
Predicted primary protein sequence characterization of PKHD-1 protein in M. musculus, H. sapiens, C. lupus familiaris
The PKHD-1 protein sequences of the three different species (M. musculus, H. sapiens, C. lupus familiaris) were retrieved from UniProt software. The details of the unique ID’s of PKHD-1 for all the three species considered for further analysis are mentioned (Table 1). UniProt is a universally acceptable database for researchers to identify a protein’s functions, taxonomy, nomenclature, subcellular location, information on post-translational modifications, their variants diseases caused by either their mutation or misfolding and details on family and domains associated with the protein [7].
The primary structure was examined and various physicochemical characters and amino acid composition were calculated using ExPasy ProtParam tool and were tabulated (Tables 2-3). The average molecular weight of PKHD1 proteins was calculated as 446386.6367 Da. The ExPasy’s ProtParam tool computes extinction coefficient for a range of (276, 278, 279, 280, 282 nm) wavelength, nevertheless, 280 nm is favored, as the thiol group of cysteine and aromatic groups of tryptophan and tyrosine in protein absorbs radiation best at 280 nm. The extinction coefficient of PKHD-1 proteins at 280 nm was 515480, 502780, 529460 M-1cm-1 in M. musculus, H. sapiens, C. lupus familiaris with respect to concentration of Cys, Trp, Tyr (Table 3). The extinction coefficient of C. lupus familiaris is comparatively high due to high concentration of Tyr (2.6%). The protein concentration and extinction coefficients aid in the quantitative study of protein-protein and protein-ligand interactions in solution [17].
The instability index value for the PKHD-1 proteins of M. musculus, H. sapiens and C. lupus familiaris were found to be 41.35, 44.73, 45.24, respectively. If the instability index is below 40, the protein is classified as stable and above 40 is classified as unstable [18]. Therefore, the PKHD-1 proteins from all three species are classified as unstable proteins. The Isoelectric Point (pI) is described as the value of pH where the charge of the protein is zero and the amino acids are in a zwitter ionic state in a protein. The pI values of M. musculus, H. sapiens and C. lupus familiaris were computed as 5.90, 6.12 and 5.95 respectively, which are less than 7, suggesting the acidic nature of PKHD-1 protein. The theoretical pI is a useful parameter for the development of buffer systems for the purification of recombinant proteins by isoelectric focusing methodology [19]. The number of negatively charged residues that is, Asp and Glu, the number of positively charged residues, that is, Arg and Lys are 379, 312 in M. musculus; 367, 313 in H. sapiens and 373, 312 in C. lupus familiaris respectively. Since the number of negatively charged residues is comparatively greater than the positively charged residues, it can be inferred that the protein is not intercellular in nature.
The half-life of PKHD-1 protein sequence of M. musculus, H. sapiens and C. lupus familiaris was found to be 30 hours in the absence of amino terminal. On the basis of this prediction, it can be inferred that the proteins were less stable in the absence of amino-terminal. The aliphatic index of a protein can be referred to as the relative volume that is occupied by aliphatic side chains, i.e., Ala, Ile, Leu, Val. It may be regarded as a positive factor for the increase of thermostability of globular proteins [20]. The aliphatic indices for the PKHD-1 were 91.62, 92.43, 93.90 for M. musculus, H. sapiens and C. lupus familiaris respectively. An inference can be drawn that the proteins are stable for a wide range of temperatures [21]. The GRAVY index values for PKHD-1 protein were -0.012, -0.020, -0.003 in M. musculus, H. sapiens and C. lupus familiaris respectively. The GRAVY index value for a peptide or protein is calculated as the sum of hydropathy values of all the amino acids, divided by the number of residues in the sequence [9,22]. The negative GRAVY values denote that the proteins are hydrophilic in nature.
The 20 amino acids were estimated using ProtParam out of which the highest percentage of amino acid is found in Leucine with 10.4, 10.0, 10.0 followed by Serine with 9.6, 9.2, 9.5 and the lowest being Tryptophan with 1.6, 1.6, 1.6 in M. musculus, H. sapiens, C. lupus familiaris respectively (Table 3).
Prediction and characterization of PKHD-1 protein secondary structures of M. musculus, H. sapiens, C. lupus familiaris
The prediction of the secondary structure of PKHD-1 proteins were evaluated using GOR tools [9]. In the designed secondary structures of PKHD-1 protein, random coils were showing 55.43, 5525, 56.55 percent in M. musculus, H. sapiens, C. lupus familiaris respectively. This is followed by Extended strands 27.10, 27.88, 25.75 and Alpha helices 17.47, 16.86, 17.70 (Table 4). Random coils aid in flexibility and conformational changes in proteins. As a result of large number of random coils, the protein is found to be extremely flexible, compact and strong bonded. These results give us a clear image that the protein is present in trans-membrane region.
| M. musculus | H. sapiens | C. lupus familiaris | ||||
|---|---|---|---|---|---|---|
| Length | Percentage (%) | Length | Percentage (%) | Length | Percentage (%) | |
| Alpha helix (Hh) | 709 | 17.47 | 687 | 16.86 | 721 | 17.7 |
| 310 helix (Gg) | 0 | 0 | 0 | 0 | 0 | 0 |
| Pi helix (Ii) | 0 | 0 | 0 | 0 | 0 | 0 |
| Beta bridge (Bb) | 0 | 0 | 0 | 0 | 0 | 0 |
| Extended strand (Ee) | 1100 | 27.1 | 1136 | 27.88 | 1049 | 25.75 |
| Beta turn (Tt) | 0 | 0 | 0 | 0 | 0 | 0 |
| Bend region (Ss) | 0 | 0 | 0 | 0 | 0 | 0 |
| Random coil (Cc) | 2250 | 55.43 | 2251 | 55.25 | 2304 | 56.55 |
| Ambiguous states | 0 | 0 | 0 | 0 | 0 | 0 |
| Other states | 0 | 0 | 0 | 0 | 0 | 0 |
Table 4: Prediction of secondary structure of PKHD-1 using GOR4 tool.
Three-dimensional modeling of PKHD-1 protein structure
The structures of PKHD-1 protein for any of the three species are unavailable in Protein Data Bank. The modeling of PKHD-1 protein was performed using SWISS-Model (Figure 1). The PKHD-1 protein in M. musculus shows 81.20% sequence identity with PKHD1 ciliary IPT domain containing fibrocystin/polyductin in Rattus norvegicus. Consecutively, the PKHD-1 protein in H. sapiens shows 80.32% sequence identity and the PKHD-1 protein in C. lupus familiaris shows 77.28% sequence identity with G8 domain-containing protein in Marmota monax. The protein modeling results for PKHD-1 using SWISS-Model is tabulated (Table 5).
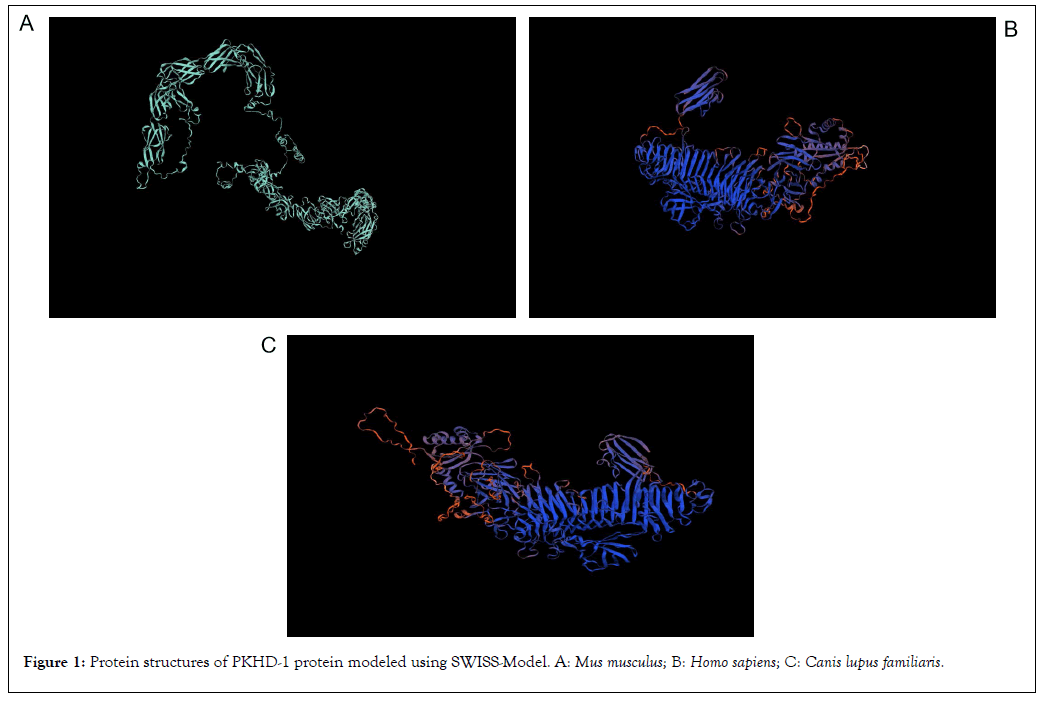
Figure 1: Protein structures of PKHD-1 protein modeled using SWISS-Model. A: Mus musculus; B: Homo sapiens; C: Canis lupus familiaris.
| S.no | Name of organism | Template organism | Template UniProt ID | Sequence identity | Sequence similarity | Coverage | Range |
|---|---|---|---|---|---|---|---|
| 1 | M. musculus | Rattus norvegicus | A0A0G2K2W1 | 81.20% | 0.55 | 0.4 | 236-1913 |
| 2 | H. sapiens | Marmota monax | A0A5E4A1X7 | 80.32% | 0.55 | 0.28 | 2636-3770 |
| 3 | C. lupus familiaris | Marmota monax | A0A5E4A1X7 | 77.28% | 0.54 | 0.28 | 2632-3765 |
Table 5: Results for protein modeling using SWISS-Model.
The ϕ and ψ distribution of Ramachandran Map generated by MolProbity server are tabulated along with summary of all-atom structure validation are evaluated for PKHD-1 protein in three species (Tables 6-7) (Figure 2). The Ramachandran outliers are defined as those amino acids with non-favourable dihedral angles and Ramachandran allowed refers to conformations where there are no steric clashes.
| S.no | Ramachandran plot calculation | Mus musculus | Homo sapiens | Canis lupus familiaris |
|---|---|---|---|---|
| 1 | Number of residues in favoured region | 91.50% | 93.80% | 93.70% |
| 2 | Number of residues in allowed region | 97.60% | 98.50% | 98.80% |
| 3 | Number of residues in outlier region | 2.40% | 1.50% | 1.20% |
Table 6: Ramachandran plot calculation using MolProbity server.
| S.no | Name of organism | Clashscore (all atoms) | Poor rotamers | Favoured rotamers | MolProbity score |
|---|---|---|---|---|---|
| 1 | Mus musculus | 1.53 (99th percentile) | 32 (2.23%) | 1357 (94.70%) | 1.66 (90th percentile) |
| 2 | Homo sapiens | 4.25 (96th percentile) | 16 (1.61%) | 958 (96.18%) | 1.77 (86th percentile) |
| 3 | Canis lupus familiaris | 3.06 (98th percentile) | 12 (1.21%) | 950 (96.15%) | 1.58 (93rd percentile) |
Table 7: All-atom structure validation using MolProbity.
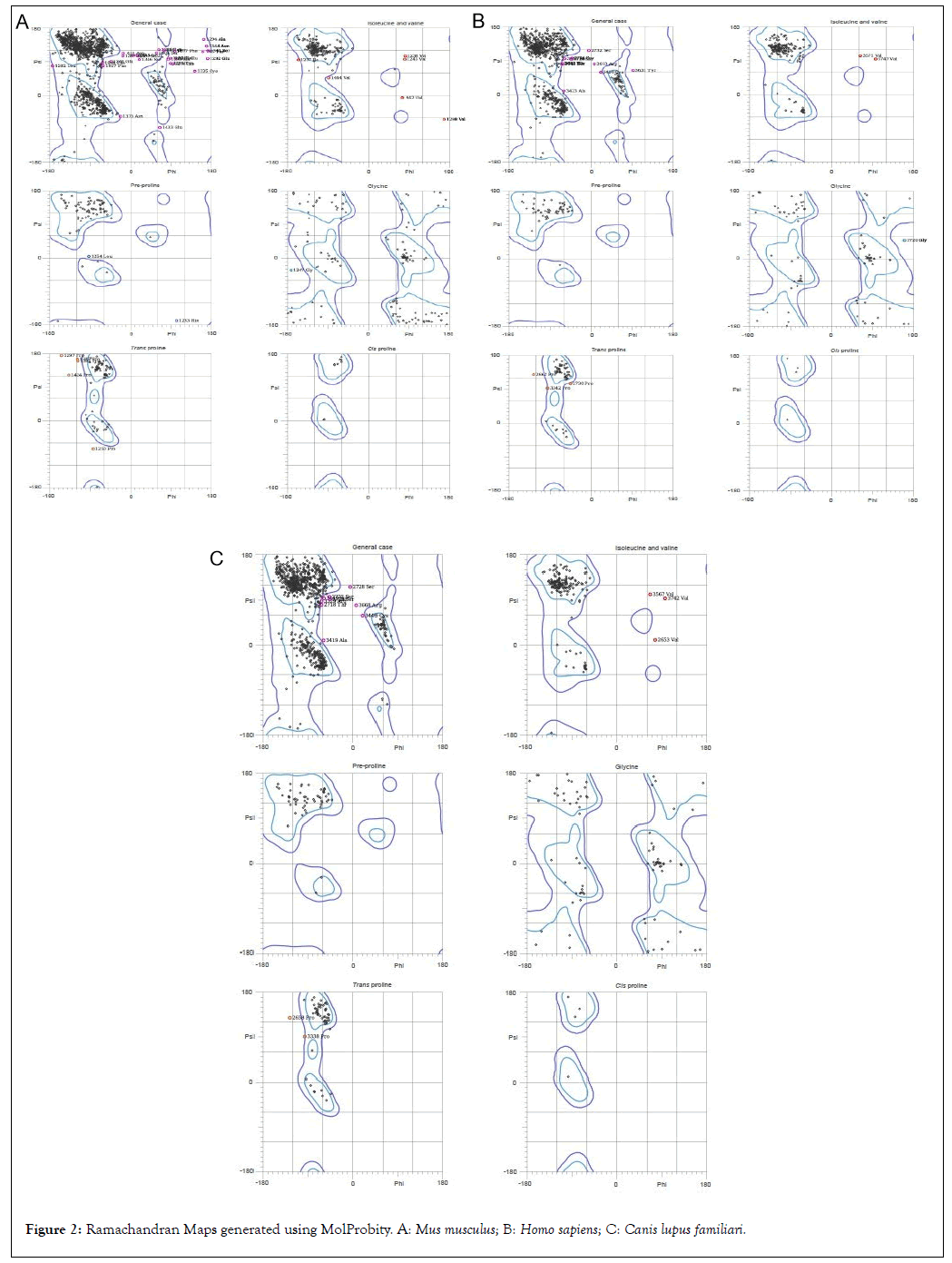
Figure 2: Ramachandran Maps generated using MolProbity. A: Mus musculus; B: Homo sapiens; C: Canis lupus familiari.
The Clashscore in MolProbity can be referred to as the number of serious steric overlaps which is greater than 0.4°A per 1000 atoms. Rotamers refer to the geometry of the amino acid side chains in a protein. Rotamer number refers to the number of those amino acids in the poor and/or favored regions along with the percentage of amino acids that come under those categories. The MolProbity score is a combination of the MolProbity clashscore, poor and favored rotamer and Ramachandran evaluations into a single quantity, standardized to lie on the same scale as that of X-ray resolution.
The protein structure after model building, was also validated through energy minimization with Z-Score using Prosa Web and quality of model was estimated using the QMEANDisCo tool from Expasy [12,23]. Z-score elucidates the variation of the total energy of the structure with regard to its energy distribution derived from random structural conformations. A more negative Z-score implies a better protein model. QMEANDisCo is a scoring function that is able to derive both for the entire structure (QMEANDisCo Global) and/or per residue (QMEANDisCo Local) absolute quality estimations based on a single model. It takes into consideration the QMEAN (Qualitative Model Energy Analysis) in addition to the distance constraints. The Z-score and QMEANDisCo Global score are tabulated (Table 8).
| S.no | Name of organism | Z-Score | QMEANDisCo Global |
|---|---|---|---|
| 1 | Mus musculus | -10.04 | 0.45 ± 0.05 |
| 2 | Homo sapiens | -11.44 | 0.55 ± 0.05 |
| 3 | Canis lupus familiaris | -11.05 | 0.53 ± 0.05 |
Table 8: Z-Scores and QMEANDisCo Global scores for overall model quality using PROSA Web and ExPasy QMEANDisCo tool.
Prediction of binding pockets
The amino acid residues constituting the binding pockets of PKHD-1 proteins in M. musculus, H. sapiens and C. lupus familiaris using P2Rank tool from PrankWeb server are tabulated (Table 9). It was observed out of the 20 polymeric pockets generated for each PKHD-1 protein in M. musculus, H. sapiens and C. lupus familiaris the first polymeric pockets were all found to be the pocket with more probability for ligand or protein attachment with probability values of 0.224 in M. musculus, 0.577 in H. sapiens and 0.592 in C. lupus familiaris.
| S.no | Name of organism | Maximum probabilty | Amino acid | Residue position |
|---|---|---|---|---|
| 1 | M. musculus | 0.224 | Thr | 1401 |
| Thr | 1441 | |||
| Arg | 1442 | |||
| Phe | 1443 | |||
| Gly | 1445 | |||
| Asp | 1446 | |||
| Gln | 1447 | |||
| Phe | 1448 | |||
| Ile | 1476 | |||
| Glu | 1478 | |||
| Thr | 1481 | |||
| Ala | 1553 | |||
| Tyr | 1555 | |||
| Cys | 1557 | |||
| 2 | H. sapiens | 0.577 | Ile | 3076 |
| Trp | 3077 | |||
| Lys | 3082 | |||
| Asn | 3084 | |||
| Gln | 3085 | |||
| Leu | 3102 | |||
| His | 3105 | |||
| His | 3131 | |||
| Tyr | 3133 | |||
| Lys | 3134 | |||
| Trp | 3255 | |||
| Trp | 3260 | |||
| 3 | C. lupus familiaris | 0.592 | Val | 3072 |
| Trp | 3073 | |||
| Lys | 3078 | |||
| Asn | 3080 | |||
| Gln | 3081 | |||
| Ile | 3098 | |||
| His | 3101 | |||
| His | 3127 | |||
| Tyr | 3129 | |||
| Lys | 3130 | |||
| Trp | 3251 | |||
| Trp | 3256 |
Table 9: Predicted binding pocket.
The comprehensive structural analysis of PKHD-1 proteins conducted in this study has profound implications for the understanding of polycystic kidney disease and related disorders. The research findings, when viewed in the context of existing research, provide valuable insights into the evolutionary and functional aspects of PKHD-1 proteins, yet several limitations must be considered. Comparing the homology models with existing data in the field, this study high- lights both the conserved regions critical for PKHD-1’s fundamental functions and the divergent domains that potentially contribute to species-specific adaptations. This comparative approach elucidates the intricate balance between evolutionary conservation and divergence in the PKHD-1 protein family. Furthermore, the identification of binding pockets in PKHD-1 proteins offers a promising avenue for targeted therapeutic interventions. Understanding these critical interaction sites provides a foundation for drug design efforts, potentially leading to novel treatments for polycystic kidney disease. Moreover, the structural insights gained from the models can guide experimental studies, informing researchers about specific regions to explore for functional characterization.
It is essential to acknowledge the limitations of this study. Firstly, the models are based on computational predictions and lack experimental validation. While state-of-the-art techniques were employed, experimental confirmation is necessary to validate the accuracy of the predicted structures and binding pockets. Secondly, the focus of this analysis was primarily on the structural aspects of PKHD-1 proteins. Functional characterization, such as enzymatic activity and protein-protein interactions, was beyond the scope of this study. Future research endeavors should bridge this gap, providing a more holistic understanding of PKHD-1 biology. Lastly, this study concentrated on a limited set of species. While Homo sapiens, Mus musculus and Canis lupus familiaris were analyzed, expanding the comparative analysis to a broader range of organisms could offer deeper insights into the evolutionary patterns of PKHD-1 proteins.
In the context of existing literature, the research findings align with previous studies that emphasize the crucial role of PKHD-1 in kidney and liver function. By expanding the structural knowledge of PKHD-1 proteins, this research contributes to the growing body of evidence that underlines the significance of this protein in health and disease.
In this study, advanced bioinformatics tools were employed to perform in-depth homology modeling and characterization of PKHD-1 in Mus musculus, Homo sapiens, Canis lupus familiaris. Through the application of SWISS-Model, ProtParam tool, GOR4 tool, PROSA Web, ExPasy QMEANDisCo tool and P2Rank tool, valuable insights into the structural aspects of PKHD-1 across different species were gained.
The primary structure along with physicochemical properties and secondary structure of PKHD-1 proteins were analyzed and evaluated using ProtParam tool and GOR4 tool respectively.
The homology model was developed using SWISS-Model Workspace from available templates in SWISS Model Template Library having the most sequence identity with the PKHD-1 sequence.
The research rigorously validated the homology models of PKHD-1 proteins using Z-Score analysis with PROSA Web and QMEANDisCo Global Score with ExPasy QMEANDisCo tool. These analyses provided a strong foundation for the reliability and accuracy of our modeled structures.
One of the significant aspects of the study involved predicting the binding pockets of PKHD-1 proteins using P2Rank tool. By identifying these binding sites, the inter-action interfaces critical for understanding the protein’s functional roles and possible therapeutic interventions can be done in the possible future.
The comparative analysis of PKHD-1 protein structures in different species not only enhances the understanding of evolutionary relationships but also lays the ground- work for future comparative functional studies. By elucidating structural similarities and differences, these findings pave the way for exploring species-specific functions and adaptations of PKHD-1.
This research significantly contributes to the fields of structural biology and biomedical research by providing detailed structural insights into PKHD-1 protein across multiple species. This comprehensive approach, combining homology modeling, validation techniques and binding pocket prediction, offers a valuable resource for researchers investigating potential PKHD-1 function, disease mechanisms and potential drug targeting strategies.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Arunannamalai SB (2024) In Silico Predictive Homology Modeling of PKHD-1 Protein: A Comparative Study among Three Different Species. J Proteomics Bioinform.17:659.
Received: 02-Feb-2024, Manuscript No. JPB-23-28035; Editor assigned: 05-Feb-2024, Pre QC No. JPB-23-28035 (PQ); Reviewed: 19-Feb-2024, QC No. JPB-23-28035; Revised: 26-Feb-2024, Manuscript No. JPB-23-28035 (R); Published: 04-Mar-2024 , DOI: 10.35248/0974-276X.24.17.659
Copyright: © 2024 Arunannamalai SB. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.