Journal of Cancer Research and Immuno-Oncology
Open Access
ISSN: 2684-1266
ISSN: 2684-1266
Research Article - (2020)Volume 6, Issue 2
Background: The X chromosome encoded FOXP3 gene is a unique regulator of the T-cell differentiation and
immunosuppressive function. The nuclear transcription factor FOXP3 gene regulates lineage-specific differentiation
in the Treg crucially maintenance of the immune homeostasis. The regulatory T-cell (Treg or CD4+ cells) play a role
in the immune response for self-antigens, allergens, and tumours. However, FOXP3 gene function is inconsistent in
tumorigenesis such as tumour-suppressive and tumour-promoting. A recent report suggested the FOXP3 gene repress
tumorigenesis per effects on proliferation and apoptosis.
Objective: My objective was to investigate the FOXP3 gene from the FOX family in between Homo sapiens and Musmusculus. The study of the FOXP3 gene is currently mandatory to explore the molecular mechanisms of the Treg
differentiation and immunosuppressive function in a particular organism.
Methods: I perform bioinformatics and computational tools and technique to the current knowledge of the FOX
family in the mammalian genome. My procedure may be useful for future functional analysis of specific gene family
in particular organisms.
Results: In this study, I conducted a compressive genome-wide survey of the FOX family in mammals. My findings
documented the FOX family play an essential role during development. The functional regulation of the FOXP3 gene
exhibits tumour suppressor activity. The specific structure, domain, motifs, phylogeny, gene expression, and
chromosome locationanalysis suggested that the FOXP3 gene is a T-cell dependent gene.
Conclusion: My analysis data concluded the FOX family plays a crucial role during development. In contrast, the
restricted expression of the FOXP3 gene in the T-cell is an immune-privileged. The ultimate function of the FOXP3
gene in tumour cells may represent a novel mechanism in the immune system.
FOXP3; FOX family; Treg or CD+ cells; T-Cell Oncogenesis
The genetic lesions of several autosomal tumour-suppressor genes intimates in the molecular pathogenesis of cancer. The cancer pathogenesis involves both inactivations of tumoursuppressor genes and activation of oncogenes. One of the most compelling aspects of cancer biology is the influence between caner-suppressor genes and oncogenes. The encoded protein of tumour-suppressor genes can inactivate oncogenes. Conversely, oncogenes may defeat the tumour-suppressor proteins. The epidemiology study suggested a role of X-linked genes controls the susceptibility of the cancers. The breakthrough of X chromosome encoded gene FOXP3 function in human leading autoimmune disorders. The FOXP3 gene function assigns critical novel insight into the biology of Treg and cellular mechanism of immune homeostasis. The immunosuppressive activity and regulatory T-cells (Treg) or CD4+ cells contribute to the progression of cancer and prevent the induction of specific immune response [1-3]. The regulatory T-cell has distinct lymphocyte lineage ability with an inhibitory property that affects the activation of the immune system. The FOXP3 gene characterizes by the specific forkhead box domain involved in the development of mammals [4,5]. The FOXP3 gene is a forkhead box DNA binding domain is conserved in evolution and consists of precise amino acid residues. The FOX (Forkheadbox) family contains 43 genes in the human genome [6]. A recent study suggested the FOXP3 gene expressed in normal breast, prostate, ovarian epithelium and also down-regulated expression in corresponding to the tumours [7-9]. The overexpression of FOXP3 gene reported in pancreatic adenocarcinoma [10], melanoma [11-13], hepatocellular carcinoma [14], leukaemia [15], bladder cancer [16], thyroid carcinoma [17] and cervical cancer [18] demonstrated critical question in the human cancer genome sequencing [19]. In contrast, the FOXP3 gene function control tumorigenesis by enabling tumour cells to lose tumour immunity. Thus previous reports supported the FOXP3 expression suppresses Treg proliferation in pancreatic carcinoma and melanoma cells [10, 13]. The FOXP3 gene expression in tumours suggested worse overall survival of the breast, bladder, and colorectal cancers [16, 20,21]. This study suggested either a pro-tumorigenic or antitumorigenic activity depend on clinical criteria. Numerous study in the animal model suggested that the inhibiting of Treg dramatically improves tumours and survival [22,23]. Treg plays a unique role in the prevention and development of effective autoimmunity. Therefore, the notable objective of this study is to evaluate the encoded FOXP3 gene that is immunogenic in nature and potential used as biomarker and target for immunotherapy. In this study, I reviewed the FOX family develops a novel therapeutics and preventives strategy in mammals. In contrast, FOXP3 gene functions play important roles in the biology of Treg and cellular mechanism of the immune homeostasis. One of the exciting developments in the cancer research findings prompted exploration of the fundamental genetic, immunogenic and molecular mechanism of the differentiation and function of Treg in oncogenesis.
Sequence and database
Primary sequence retrieves from the different specific database (UniProt, KEGG, GenBank, EMBL, DDBJ and NCBI), and performed web base application SMART for identification of the particular domain in the query sequence. Pfam searched for retrieving protein family information. SWISS-MODEL performs for the structure prediction. The SWISS-MODEL is structural bioinformatics web-server for comparative modelling of 3D structure. The most accurate method for generating rational 3D structure and routinely used in many practical applications. This method makes the experimental protein structure to a build model of evolutionarily related proteins. The SWISS-MODEL is a continuously updated database of homology or comparative model of the organism proteome for biomedical research. Genome: The genome sequences download from genomic data in different specialized databases (NCBI and Ensemble).
Standalone tools and GO annotation
HMMER executes using multiple sequence alignments of the specific domain as a profile search. HMMER is a statistical algorithm, making multiple sequence alignment (MSA) of the particular domain as a profile search and implemented methods using probabilistic models called the profile hidden Markov model. The standalone BLAST performs for homologs gene in selected organisms. The BLAST2GO performs for the gene ontology annotation, is a bioinformatics and computational tool for high-throughput gene ontology annotation of the particular sequence. The practical information of the genes retrieves via Gene Ontology (GO) annotation, a controlled vocabulary of the functional attribute.
Domain, motif, and phylogeny
Multiple sequence alignment (MSA) method for calculate the best match of the homologs sequences and line them up so identities, similarities, and differences can observe. MSA of highest hits sequence analysis carried out by web-based tool MultAlin for identification of conserved domain. The identification of the molecular evolutionary relationship of the specific gene in the Homo sapiens and Musmusculus, MEGA7 performed for constructing a phylogenetic tree using Neighbor- Joining Methods. The MEME suite retrieves for the sequence motifs is a computational web-based tool for discovery and analysis of specific motifs.
Gene expression and chromosome location
The expression analysis carried out using the GENEVESTIGATOR tool is a high-performance search engine of gene expression in different biological contexts. GENEVESTIGATOR use to identify and validate novel targets. Chromosome location retrieves using gene card is a database of the human genes provide genomic information of all known and predicted human genes. This database is currently available for biomedical research such as gene, encoded protein and associated disease.
Structural Analysis
The primary sequence demonstrated the specific composition of nucleotides and peptides. The sequence composed of 1296 nucleotides and 431 peptides with 83 peptides binding to the DNA sequence is well-known as a forkhead domain (Table 1a and 1b). The peptide structure illustrated the forkhead domain involved in DNA binding. The forkhead domain also is known as “ winged helix ” domain. The 3D structure demonstrated alpha and beta proteins binds with B-DNA as a monomer interacts with the DNA backbone. The alpha helices assume a compact structure that presents in the third helix to the major grove. The protein comprises a twisted antiparallel beta structure and random coil that interact with the minor groove (Figure 1).
| >FOXP3 ATGCCCAACCCCAGGCCTGGCAAGCCCTCGGCCCCTTCCTTGGCCCTTGGCCCATCCCCAGGAGCCTCGCCCAGCTGGAGGGCTGCACCCAAAGCCTCAGACCTGCTGGGGGCCCGGGG CCCAGGGGGAACCTTCCAGGGCCGAGATCTTCGAGGCGGGGCCCATGCCTCCTCTTCTTCCTTGAACCCCATGCCACCATCGCAGCTGCAGCTCTCAACGGTGGATGCCCACGCC CGGACCCCTGTGCTGCAGGTGCACCCCCTGGAGAGCCCAGCCATGATCAGCCTCACACCACCCACCACCGCCACTGGGGTCTTCTCCCTCAAGGCCCGGCCTGGCCTCCCACCTG GGATCAACGTGGCCAGCCTGGAATGGGTGTCCAGGGAGCCGGCACTGCTCTGCACCTTCCCAAATCCCAGTGCACCCAGGAAGGACAGCACCCTTTCGGCTGTGCCCCAGAGCTC CTACCCACTGCTGGCAAATGGTGTCTGCAAGTGGCCCGGATGTGAGAAGGTCTTCGAAGAGCCAGAGGACTTCCTCAAGCACTGCCAGGCGGACCATCTTCTGGATGAGAAGGGC AGGGCACAATGTCTCCTCCAGAGAGAGATGGTACAGTCTCTGGAGCAGCAGCTGGTGCTGGAGAAGGAGAAGCTGAGTGCCATGCAGGCCCACCTGGCTGGGAAAATGGCACTGA CCAAGGCTTCATCTGTGGCATCATCCGACAAGGGCTCCTGCTGCATCGTAGCTGCTGGCAGCCAAGGCCCTGTCGTCCCAGCCTGGTCTGGCCCCCGGGAGGCCCCTGACAGCCT GTTTGCTGTCCGGAGGCACCTGTGGGGTAGCCATGGAAACAGCACATTCCCAGAGTTCCTCCACAACATGGACTACTTCAAGTTCCACAACATGCGACCCCCTTTCACCTACGCC ACGCTCATCCGCTGGGCCATCCTGGAGGCTCCAGAGAAGCAGCGGACACTCAATGAGATCTACCACTGGTTCACACGCATGTTTGCCTTCTTCAGAAACCATCCTGCCACCTGGA AGAACGCCATCCGCCACAACCTGAGTCTGCACAAGTGCTTTGTGCGGGTGGAGAGCGAGAAGGGGGCTGTGTGGACCGTGGATGAGCTGGAGTTCCGCAAGAAACGGAGCCAGAG GCCCAGCAGGTGTTCCAACCCTACACCTGGCCCCTGA |
Table 1 (a): Primary Structure –Nucleotide.
| >FOXP3 MPNPRPGKPSAPSLALGPSPGASPSWRAAPKASDLLGARGPGGTFQGRDLRGGAHASSSSLNPMPPSQLQLPTLPLVMVAPSGARLGPLPHLQALLQDRPHFMHQLSTVDAHARTPVLQ VHPLESPAMISLTPPTTATGVFSLKARPGLPPGINVASLEWVSREPALLCTFPNPSAPRKDSTLSAVPQSSYPLLANGVCKWPGCEKVFEEPEDFLKHCQADHLLDEKGRAQCLL QREMVQSLEQQLVLEKEKLSAMQAHLAGKMALTKASSVASSDKGSCCIVAAGSQGPVVPAWSGPREAPDSLFAVRRHLWGSHGNSTFPEFLHNMDYFKFHNMRPPFTYATLIRWA ILEAPEKQRTLNEIYHWFTRMFAFFRNHPATWKNAIRHNLSLHKCFVRVESEKGAVWTVDELEFRKKRSQRPSRCSNPTPGP |
Table 1 (b): Primary Structure -Peptide.
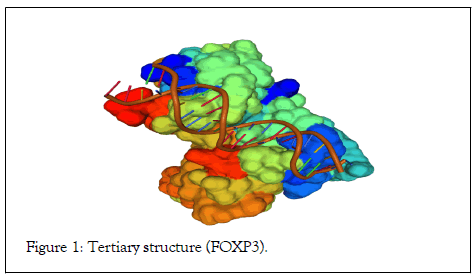
Figure 1: Tertiary structure (FOXP3).
Genome-wide Analysis
The genome-wide analysis by HMMER algorithm results shown that the multiple hits of 114 and 88 of the particular forkhead domains in Homo sapiens and Musmusculus respectively (Table 2). The standalone BLAST results represent 116 and 88 of homologs in Homo sapiens and Musmusculus, respectively (Table 2). Multiple hits selected from both organisms for gene ontology (GO) annotation (Table 3). The gene ontology annotation confirms that a total of 6 and 4 of the FOXP3 gene in Homo sapiens and Musmusculus respectively (Table 2).
| Organisms | HMMER Hits | BLAST Hits |
|---|---|---|
| Homo sapiens | 114 | 116 |
| Musmusculus | 88 | 88 |
| Total | 202 | 204 |
| Summary of the Fork head domain | ||
| Gene | Homo sapiens | Musmusculus |
| FOXP3 | 6 | 4 |
| FOXP1 | 11 | 13 |
| FOXP4 | 5 | 4 |
| FOXP2 | 11 | 4 |
| FOXJ3 | 7 | 3 |
| FOXJ2 | 2 | 5 |
| FOXC2 | 1 | 1 |
| FOXJ1 | 1 | 1 |
| FOXC1 | 1 | 1 |
| FOXN3 | 10 | 5 |
| FOXL1 | 3 | 2 |
| FOXG1 | 1 | 2 |
| FOXE3 | 1 | 1 |
| FOXK2 | 4 | 1 |
| FOXE1 | 1 | 1 |
| FOXI3 | 1 | 2 |
| FOXI1 | 2 | 1 |
| FOXN2 | 3 | 2 |
| FOXA1 | 1 | 1 |
| FOXA2 | 2 | 3 |
| FOXA3 | 1 | 1 |
| FOXS1 | 1 | 1 |
| FOXI2 | 1 | 1 |
| FOXF1 | 1 | 1 |
| FOXD1 | 1 | 2 |
| FOXD2 | 1 | 0 |
| FOXF2 | 1 | 1 |
| FOXL2 | 1 | 1 |
| FOXD3 | 1 | 1 |
| FOXM1 | 5 | 4 |
| FOXO6 | 2 | 1 |
| FOXD4 | 6 | 1 |
| FOXO4 | 2 | 2 |
| FOXK1 | 1 | 1 |
| FOXO3 | 2 | 4 |
| FOXO1 | 1 | 1 |
| FOXN1 | 2 | 1 |
| FOXQ1 | 1 | 1 |
| FOXB1 | 1 | 1 |
| FOXB2 | 1 | 1 |
| FOXN4 | 3 | 1 |
| FOXH1 | 1 | 1 |
| FOXR1 | 2 | 0 |
| FOXR2 | 1 | 2 |
| FOXD5 | 0 | 0 |
| FOXD6 | 0 | 0 |
| FOXE2 | 0 | 0 |
| Total | 114 | 87 |
Table 2: Summary of the (a) Forkhead domain and (b) FOX family
| Gene Id | Gene | Protein |
|---|---|---|
| ENSP00000365372.2 | FOXP3 | Forkhead box P3 |
| ENSP00000428952.1 | FOXP3 | Forkhead box P3 |
| ENSP00000365380.4 | FOXP3 | forkhead box protein P3 isoform X3 |
| ENSP00000396415.3 | FOXP3 | forkhead box protein P3 isoform X1 |
| ENSP00000365369.1 | FOXP3 | forkhead box protein P3 isoform X1 |
| ENSP00000451208.1 | FOXP3 | forkhead box protein P3 isoform X1 |
| ENSP00000418225.1 | FOXP1 | forkhead box protein P1 isoform X1 |
| ENSP00000482847.1 | FOXP1 | forkhead box protein P1 isoform X1 |
| ENSP00000333560.4 | FOXP1 | forkhead box protein P1 isoform X4 |
| ENSP00000417857.1 | FOXP1 | forkhead box protein P1 isoform X2 |
| ENSP00000420736.1 | FOXP1 | forkhead box protein P1 isoform X4 |
| ENSP00000418524.1 | FOXP1 | forkhead box protein P1 isoform X3 |
| ENSP00000318902.4 | FOXP1 | forkhead box protein P1 isoform X4 |
| ENSP00000418102.1 | FOXP1 | forkhead box protein P1 isoform X4 |
| ENSP00000419393.1 | FOXP1 | forkhead box protein P1 isoform X4 |
| ENSP00000362154.3 | FOXP4 | forkhead box protein P4 isoform X5 |
| ENSP00000386958.1 | FOXP4 | forkhead box protein P4 isoform X4 |
| ENSP00000362148.3 | FOXP4 | forkhead box protein P4 isoform X2 |
| ENSP00000309823.4 | FOXP4 | forkhead box protein P4 isoform X1 |
| ENSP00000362151.1 | FOXP4 | forkhead box protein P4 isoform X1 |
| ENSP00000488944.1 | FOXP2 | forkhead box protein P2 isoform X3 |
| ENSP00000377129.3 | FOXP2 | forkhead box protein P2 |
| ENSP00000385069.4 | FOXP2 | forkhead box protein P2 |
| ENSP00000377135.2 | FOXP2 | forkhead box protein P2 |
| ENSP00000489135.1 | FOXP2 | forkhead box protein P2 |
| ENSP00000386200.3 | FOXP2 | forkhead box protein P2 |
| ENSP00000265436.7 | FOXP2 | forkhead box protein P2 |
| ENSP00000377132.2 | FOXP2 | forkhead box protein P2 |
| ENSP00000489073.1 | FOXP2 | forkhead box protein P2 |
| ENSP00000489229.1 | FOXP2 | forkhead box protein P2 |
| ENSP00000484803.1 | FOXP1 | forkhead box protein P1 isoform X1 |
| ENSP00000377130.3 | FOXP2 | forkhead box protein P2 |
| ENSP00000418883.1 | FOXP1 | forkhead box protein P1 isoform X4 |
| ENSP00000403060.1 | FOXJ3 | forkhead box protein J3-like |
| ENSP00000393408.1 | FOXJ3 | forkhead box protein J3 isoform X3 |
| ENSP00000354449.1 | FOXJ3 | forkhead box protein J3 isoform X3 |
| ENSP00000354620.1 | FOXJ3 | forkhead box protein J3 isoform X1 |
| ENSP00000361653.1 | FOXJ3 | forkhead box protein J3 isoform X1 |
| ENSP00000361654.1 | FOXJ3 | forkhead box protein J3 isoform X1 |
| ENSP00000439044.1 | FOXJ3 | forkhead box protein J3 isoform X1 |
| ENSP00000403411.2 | FOXJ2 | forkhead box protein J2 |
| ENSP00000162391.3 | FOXJ2 | forkhead box protein J2 isoform X1 |
| ENSP00000326371.4 | FOXC2 | forkhead box protein C2 |
| ENSP00000323880.4 | FOXJ1 | forkhead box protein J1 |
| ENSP00000370256.2 | FOXC1 | forkhead box protein C1 |
| ENSP00000451135.1 | FOXN3 | forkhead box protein N3 isoform X3 |
| ENSP00000326272.3 | FOXL1 | forkhead box protein L1 |
| ENSP00000339004.3 | FOXG1 | forkhead box protein G1 |
| ENSP00000334472.2 | FOXE3 | forkhead box protein E3 |
| ENSP00000452005.1 | FOXN3 | forkhead box protein N3 isoform X1 |
| ENSP00000452227.1 | FOXN3 | forkhead box protein N3 isoform X1 |
| ENSP00000479114.1 | FOXN3 | forkhead box protein N3 isoform X1 |
| ENSP00000261302.5 | FOXN3 | forkhead box protein N3 isoform X1 |
| ENSP00000343288.4 | FOXN3 | forkhead box protein N3 isoform X1 |
| ENSP00000472376.1 | FOXL1 | forkhead box protein L1 |
| ENSP00000433167.1 | FOXK2 | forkhead box protein K2 |
| ENSP00000364265.3 | FOXE1 | forkhead box protein E1 |
| ENSP00000478384.2 | FOXI3 | forkhead box protein I3 |
| ENSP00000304286.5 | FOXI1 | forkhead box I1 |
| ENSP00000432663.2 | FOXK2 | forkhead box protein K2 |
| ENSP00000388486.1 | FOXN2 | forkhead box protein N2 |
| ENSP00000250448.2 | FOXA1 | hepatocyte nuclear factor 3-alpha |
| ENSP00000484534.1 | FOXN2 | forkhead box protein N2 |
| ENSP00000343633.3 | FOXN2 | forkhead box protein N2 |
| ENSP00000304004.1 | FOXA3 | hepatocyte nuclear factor 3-gamma |
| ENSP00000436108.2 | FOXK2 | forkhead box protein K2 |
| ENSP00000366319.4 | FOXA2 | hepatocyte nuclear factor 3-beta isoform X2 |
| ENSP00000335677.5 | FOXK2 | forkhead box protein K2 |
| ENSP00000400341.3 | FOXA2 | hepatocyte nuclear factor 3-beta isoform X1 |
| ENSP00000365145.3 | FOXS1 | forkhead box protein S1 |
| ENSP00000373572.4 | FOXI2 | forkhead box protein I2 |
| ENSP00000262426.4 | FOXF1 | forkhead box protein F1 |
| ENSP00000481581.1 | FOXD1 | forkhead box protein D1 |
| ENSP00000335493.5 | FOXD2 | forkhead box protein D2 |
| ENSP00000259806.1 | FOXF2 | forkhead box protein F2 |
| ENSP00000333188.3 | FOXL2 | forkhead box protein L2 |
| ENSP00000415483.2 | FOXI1 | forkhead box protein I1 isoform X2 |
| ENSP00000360157.2 | FOXD3 | forkhead box protein D3 |
| ENSP00000354492.3 | FOXM1 | Forkhead box M1 |
| ENSP00000486536.1 | FOXM1 | Forkhead box M1 |
| ENSP00000352901.3 | FOXM1 | forkhead box protein M1 isoform X1 |
| ENSP00000342307.2 | FOXM1 | forkhead box protein M1 isoform X1 |
| ENSP00000492766.1 | FOXL1 | forkhead box L1-like |
| ENSP00000493184.1 | FOXO6 | forkhead box protein O6 |
| ENSP00000486069.5 | FOXO6 | forkhead box protein O6 |
| ENSP00000302756.5 | FOXD4 | forkhead box protein D4-like 1 |
| ENSP00000363377.3 | FOXO4 | forkhead box protein O4 |
| ENSP00000328720.4 | FOXK1 | forkhead box protein K1 |
| ENSP00000484875.1 | FOXD4 | Forkhead box D4-like 6 |
| ENSP00000341961.2 | FOXD4 | Forkhead box D4-like 6 |
| ENSP00000366637.1 | FOXD4 | Forkhead box D4-like 6 |
| ENSP00000366630.1 | FOXD4 | Forkhead box D4-like 6 |
| ENSP00000339527.6 | FOXO3 | forkhead box protein O3 |
| ENSP00000385824.1 | FOXO3 | forkhead box protein O3 |
| ENSP00000371940.2 | FOXD4 | Forkhead box D4 |
| ENSP00000368880.4 | FOXO1 | forkhead box protein O1 |
| ENSP00000226247.2 | FOXN1 | forkhead box protein N1 |
| ENSP00000464645.1 | FOXN1 | forkhead box protein N1 |
| ENSP00000296839.2 | FOXQ1 | forkhead box protein Q1 |
| ENSP00000379369.4 | FOXB1 | forkhead box protein B1 |
| ENSP00000342209.3 | FOXO4 | forkhead box protein O4 isoform X2 |
| ENSP00000365898.1 | FOXB2 | forkhead box protein B2 |
| ENSP00000299162.5 | FOXN4 | forkhead box protein N4 |
| ENSP00000366534.4 | FOXH1 | forkhead box protein H1 |
| ENSP00000450684.1 | FOXN3 | forkhead box protein N3 isoform X1 |
| ENSP00000347354.1 | FOXN4 | forkhead box protein N4 |
| ENSP00000474754.1 | FOXN4 | forkhead box protein N4 |
| ENSP00000451024.1 | FOXN3 | forkhead box protein N3 isoform X3 |
| ENSP00000314806.3 | FOXR1 | forkhead box protein R1 |
| ENSP00000487495.3 | FOXR1 | forkhead box protein R1 |
| ENSP00000450833.1 | FOXN3 | forkhead box protein N3 isoform X1 |
| ENSP00000451437.1 | FOXN3 | forkhead box protein N3 isoform X3 |
| ENSP00000442309.1 | FOXM1 | forkhead box protein M1 isoform X1 |
| ENSP00000427329.2 | FOXR2 | forkhead box protein R2 |
| Homo sapiens | ||
| Gene Id | Gene | Protein |
| ENSMUSP00000041953.6 | FOXP3 | forkhead box protein P3 |
| ENSMUSP00000111403.1 | FOXP3 | forkhead box protein P3 |
| ENSMUSP00000111404.1 | FOXP3 | forkhead box protein P3 |
| ENSMUSP00000111405.1 | FOXP3 | forkhead box protein P3 |
| ENSMUSP00000135809.1 | FOXP1 | forkhead box protein P1 isoform X1 |
| ENSMUSP00000134817.1 | FOXP1 | forkhead box protein P1 isoform X1 |
| ENSMUSP00000135098.1 | FOXP1 | forkhead box protein P1 isoform X1 |
| ENSMUSP00000135635.1 | FOXP1 | forkhead box protein P1 isoform X1 |
| ENSMUSP00000135041.1 | FOXP1 | forkhead box protein P1 isoform X1 |
| ENSMUSP00000108952.2 | FOXP1 | forkhead box protein P1 isoform X1 |
| ENSMUSP00000135181.1 | FOXP1 | forkhead box protein P1 isoform X1 |
| ENSMUSP00000135764.1 | FOXP1 | forkhead box protein P1 isoform X1 |
| ENSMUSP00000108954.2 | FOXP1 | forkhead box protein P1 isoform X1 |
| ENSMUSP00000073953.5 | FOXP1 | forkhead box protein P1 isoform X1 |
| ENSMUSP00000108948.2 | FOXP1 | forkhead box protein P1 isoform X1 |
| ENSMUSP00000108950.2 | FOXP1 | forkhead box protein P1 isoform X1 |
| ENSMUSP00000108890.1 | FOXP4 | forkhead box protein P4 isoform X4 |
| ENSMUSP00000108887.1 | FOXP4 | forkhead box protein P4 isoform X1 |
| ENSMUSP00000108888.1 | FOXP4 | forkhead box protein P4 isoform X1 |
| ENSMUSP00000094916.2 | FOXP4 | forkhead box protein P4 isoform X1 |
| ENSMUSP00000111134.1 | FOXP2 | forkhead box protein P2 isoform X4 |
| ENSMUSP00000111132.1 | FOXP2 | forkhead box protein P2 isoform X4 |
| ENSMUSP00000031545.7 | FOXP2 | forkhead box protein P2 isoform X4 |
| ENSMUSP00000111137.1 | FOXP2 | forkhead box protein P2 isoform X4 |
| ENSMUSP00000108955.3 | FOXP1 | forkhead box protein P1 isoform X5 |
| ENSMUSP00000145438.1 | FOXJ2 | forkhead box protein J2 |
| ENSMUSP00000123815.1 | FOXJ3 | forkhead box protein J3 isoform X3 |
| ENSMUSP00000124806.1 | FOXJ3 | forkhead box protein J3 isoform X3 |
| ENSMUSP00000101917.2 | FOXJ3 | forkhead box protein J3 isoform X2 |
| ENSMUSP00000035746.8 | FOXJ3 | forkhead box protein J3 isoform X1 |
| ENSMUSP00000003238.7 | FOXJ2 | forkhead box protein J2 |
| ENSMUSP00000137645.1 | FOXJ2 | forkhead box protein J2 |
| ENSMUSP00000055290.6 | FOXC2 | forkhead box protein C2 |
| ENSMUSP00000038351.6 | FOXJ1 | forkhead box protein J1 |
| ENSMUSP00000052196.2 | FOXC1 | forkhead box protein C1 |
| ENSMUSP00000137732.1 | FOXL1 | forkhead box protein L1 |
| ENSMUSP00000050445.2 | FOXE3 | forkhead box protein E3 |
| ENSMUSP00000021333.3 | FOXG1 | forkhead box protein G1 |
| ENSMUSP00000136372.1 | FOXG1 | forkhead box protein G1 |
| ENSMUSP00000036035.4 | FOXN3 | forkhead box protein N3 isoform X3 |
| ENSMUSP00000082189.1 | FOXN3 | forkhead box protein N3 isoform X3 |
| ENSMUSP00000135082.1 | FOXN3 | forkhead box protein N3 isoform X3 |
| ENSMUSP00000135749.2 | FOXN3 | forkhead box protein N3 |
| ENSMUSP00000135814.1 | FOXN3 | forkhead box protein N3 |
| ENSMUSP00000092715.2 | FOXE1 | forkhead box protein E1 |
| ENSMUSP00000065664.5 | FOXI3 | forkhead box protein I3 |
| ENSMUSP00000125380.1 | FOXI3 | forkhead box protein I3 |
| ENSMUSP00000041118.6 | FOXA1 | hepatocyte nuclear factor 3-alpha |
| ENSMUSP00000145473.1 | FOXM1 | forkhead box protein M1 isoform X1 |
| ENSMUSP00000043173.5 | FOXA3 | hepatocyte nuclear factor 3-gamma |
| ENSMUSP00000107857.2 | FOXN2 | forkhead box protein N2 |
| ENSMUSP00000100725.3 | FOXD1 | forkhead box protein D1 |
| ENSMUSP00000058651.2 | FOXI1 | forkhead box protein I1 |
| ENSMUSP00000101719.1 | FOXK2 | forkhead box protein K2 |
| ENSMUSP00000045918.3 | FOXA2 | hepatocyte nuclear factor 3-beta isoform X2 |
| ENSMUSP00000105590.1 | FOXA2 | hepatocyte nuclear factor 3-beta isoform X1 |
| ENSMUSP00000096806.2 | FOXS1 | forkhead box protein S1 |
| ENSMUSP00000137662.1 | FOXF1 | forkhead box protein F1 |
| ENSMUSP00000066868.3 | FOXD1 | forkhead box protein D1 |
| ENSMUSP00000053641.5 | FOXI2 | forkhead box protein I2 |
| ENSMUSP00000046789.2 | FOXF2 | forkhead box protein F2 |
| ENSMUSP00000053297.2 | FOXL2 | forkhead box protein L2 |
| ENSMUSP00000118378.1 | FOXN2 | forkhead box protein N2 |
| ENSMUSP00000145305.1 | FOXM1 | forkhead box protein M1 isoform X1 |
| ENSMUSP00000084541.3 | FOXD3 | forkhead box protein D3 |
| ENSMUSP00000107776.1 | FOXM1 | Forkhead box M1 |
| ENSMUSP00000073041.6 | FOXM1 | forkhead box protein M1 |
| ENSMUSP00000137099.1 | FOXL1 | forkhead box L1-like |
| ENSMUSP00000099716.3 | FOXO6 | forkhead box protein O6 |
| ENSMUSP00000058575.2 | FOXD4 | forkhead box protein D4 |
| ENSMUSP00000059420.4 | FOXO4 | forkhead box protein O4 |
| ENSMUSP00000050683.3 | FOXO3 | forkhead box protein O3 |
| ENSMUSP00000101141.1 | FOXO3 | forkhead box protein O3 |
| ENSMUSP00000135380.1 | FOXO3 | forkhead box protein O3 |
| ENSMUSP00000072616.5 | FOXK1 | forkhead box protein K1 |
| ENSMUSP00000055308.5 | FOXO1 | forkhead box protein O1 |
| ENSMUSP00000103929.1 | FOXN1 | forkhead box protein N1 isoform X1 |
| ENSMUSP00000036952.8 | FOXQ1 | forkhead box protein Q1 |
| ENSMUSP00000096197.2 | FOXB1 | forkhead box protein B1 |
| ENSMUSP00000116165.1 | FOXO4 | forkhead box protein O4 |
| ENSMUSP00000072687.2 | FOXB2 | forkhead box protein B2 |
| ENSMUSP00000047951.5 | FOXN4 | forkhead box protein N4 |
| ENSMUSP00000036591.4 | FOXH1 | forkhead box protein H1 |
| ENSMUSP00000093984.3 | FOXR2 | forkhead box protein R2 |
| ENSMUSP00000128452.1 | FOXR2 | forkhead box protein R2 |
| ENSMUSP00000101140.1 | FOXO3 | forkhead box protein O3 |
| ENSMUSP00000123923.1 | FOXJ3 | forkhead box protein J3 isoform X4 |
| ENSMUSP00000134081.1 | FOXA2 | hepatocyte nuclear factor 3-beta isoform X2 |
| Musmusculus | ||
Table 3: Summary of the Gene Ontology annotation (a) Homo sapiens and (b) Musmusculus.
Analysis of the domain, motifs, and phylogeny
The highest hits of the FOXP3 gene listed from both organisms for sequence alignment, a multiple sequence alignment (MSA) determine the conserved domain (Figure 2). The high consensus (90%) sequence indicates the extended forkhead domain and their specific motifs (Figure 3a, 3b, and 3c). The phylogenetic tree demonstrated the molecular evolutionary relationship of the FOXP3 gene in between Homo sapiens and Musmusculus. Particular clades represent the multifunctional forkhead domain involved genes in both organisms (Figure 4).
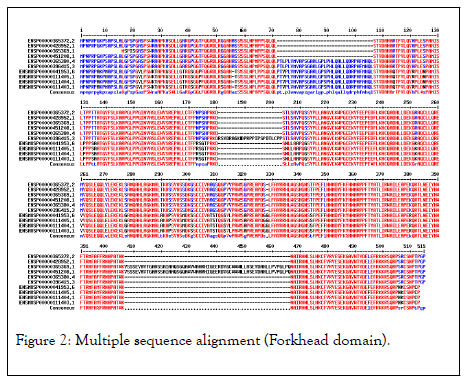
Figure 2: Multiple sequence alignment (Forkhead domain).

Figure 3: Sequence motifs (a, b, and c).
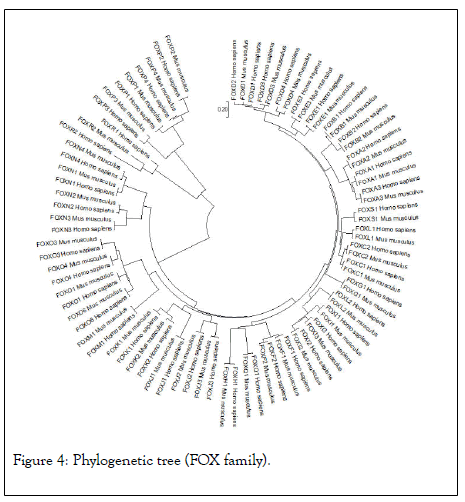
Figure 4: Phylogenetic tree (FOX family).
Analysis of the gene expression and chromosome location
The gene expression analysis of four anatomical cancer categories shown that the low level of FOXP3 expression and express in neoplasm of secondary/ill-defined, malignant neoplasm, malignant melanoma (unstable behavior) is shown in Figure 5. The chromosome location study demonstrated that the FOXP3 located band Xp11.23, start 49,250,436 bp and end 49,270,477 bp is shown in Figure 6.
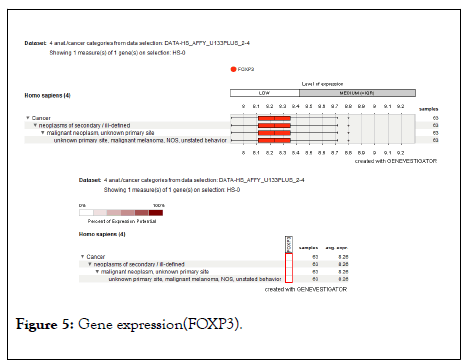
Figure 5: Gene expression(FOXP3).
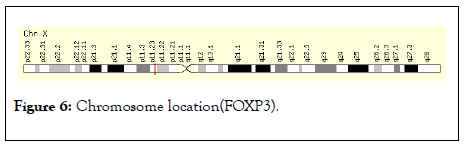
Figure 6: Chromosome location(FOXP3).
The immune system is a subject of the self and no selfdiscrimination both is directly fighting against the pathogens and maintains tolerance to self-antigens in mammals. Immune tolerance of the T-cells is achieving through central and peripheral strategies. The identification and characterization of regulatory T-cells (Treg or CD4+ cells) play unique roles in immune tolerance to self and transplant antigens [24-28].
Since the T-cell deficiency is a subject of lack of Treg in individual, it remains unknown the Treg function in the adult develop the immune system is not critical in the newborn. According to the genome, sequencing study suggested the elimination of Treg or CD4+ cells in the adult’s restively mild immune-mediated lesions and compression associated with congenital T-cell deficiency. In this study, my findings suggested the fundamental roles of FOXP3 gene dependent for differentiation and Treg function in the immune system and raise a question, how numerous inherent mechanisms prevent differentiation of Treg in the thymus and restrain their activity in the periphery to ensure self-tolerance.
FOXP3 gene considers as a master regulatory function in typically derived naturally occurring Treg (CD4+ cells). The FOXP3 express in Treg cells found in the major histocompatibility complex (MHC) class 2 restricted CD4+ cells and express high levels of CD25 (IL-2). An addition of FOXP3 also express CD4+ cells and CD25+ cells appears in minor histocompatibility complex (MHC) class 1 restricted CD8+ cells expression in Treg [29,30].
Therefore, the expression of CD8+ cells can kill tumour cells by the release of perforin and granzymes. The Treg also included Type 1 regulatory cells (Th1 or CD4+ cells), T helper 3 cells (Th3), CD8, CD28, and HLA-E restricted T-cells. The Treg function is associated with CD28 and CTLA-4 receptor molecules both potentially bind with two natural ligands such as CD80 and CD86 are capable of Treg-suppressive capacity [31].
The CD86 (B7-2) strongly enhances suppression by CD4+ CD25+ and Treg cells, the blocking of CD80 (B7-1) enhance proliferative responses by reducing Treg suppression. Comparative expression of CD80 and CD86 on dendritic cells regulates during the progression of abnormal to a normal state with the ability of Treg-suppressive responses. The CD80 and CD86 have rival functions through CD28 and CD152 (CTLA-4) on Treg that has remarkable effect for control of immune responses. The initiation of Treg cells and expression of immune checkpoint molecules includes CTLA-4, PD-1 (CD279), TIM-3, LAG-3, and TIGIT [32,33].
The prominent rule of PD-1 receptor is mainly express on T cells and interacts with their two ligands PD-L1 (B7-H1, CD274) and PD-L2 (B7-DC, CD273) play roles in the initiation and maintenance of peripheral tolerance mechanism [34]. The PDL1 and PD-L2 express on antigen-presenting cells (APCs) and tumours release a negative signal to T cells, which is called T-cell exhaustion. Antibodies binding CTLA-4, PD-1 and PD-L1 have striking ability to enhance anti-cancer immunotherapy [35,36].
Therefore, Treg activity can evoke anti-tumour immunity and acknowledge inhibition of the T-cells function. The peptide inhibiter protein probably binds to the forkhead/winged-helix transcription factor 3 (FOXP3) which necessary for the development and function of Treg (CD4+ cells). The CD4+ cells produce transforming growth factor-beta and IL10 for suppressive function in the immune system and allow cell survival. The peptide P60 enter into the cells and inhibits FOXP3 nuclear translocation and reduces its ability to suppress the transcription factors NF-kB and NFAT both regulate transcription of DNA, cytokine production and cell survival. The functional evidence of the FOXP3 gene represses the expression of CD340, SKP2, SATB1 and MYC oncogenes and induces expression of tumour suppressor genes P21 and LATS2. The FOXP3 gene also pioneers of the anti-tumourenzymes such as CD39 and CD8 [37].
The functional inhibition of Treg by the FOXP3-inhibitory peptide initiates a strategy to enhance antitumor immunotherapies [3]. This study describes the significant advances of the FOXP3 gene as an X-linked tumour suppressor gene in between Homo sapiens and Musmusculus. This work represents a compelling exception and widely accepted theory of the inactivation of tumour suppressor genes. The oncogenes are a heterogeneous group that is generally expressed in the cells and trophoblast and also aberrantly activated in various cancers [38].
A subset of oncogenes encodes antigens that are immunogenic and elicit humoral and cellular immune responses in cancers [39]. Limited evidence supported the FOXP3 gene is a tumour suppressor gene. In this report, my finding suggested the expression of FOXP3 gene in cancer cells provide evidence that this could be important in tumour escape mechanisms. In this study, I discuss the multiple in-silico analysis and comprehensive genome-wide survey of the FOX family is involved in the development process in mammals. In contrast, the restricted expression of FOXP3 gene in the genome is an immuneprivileged and therapeutic strategy in cancers. The conserved domain, motifs, phylogeny, gene expression, and chromosome location analysis suggested the mechanism of the FOXP3 gene is conserved in evolution. The phylogeny analysis has apparent the large gene family consist of specific genes is closely related to each other.
Thus study suggested the FOXP3 gene acts as transcriptional activator and repressor and also targets makeup as a T-cell dependent gene. The fundamental contribution of the FOXP3 gene in tumour cells may represent a novel mechanism in the immune system. This study is consistent with the results of an in-silico analysis of FOXP3 gene target in T-cells. Therefore, my finding supported the genetic, immunologic and molecular mechanisms of the X chromosome encoded FOXP3 gene associated with T-cell oncogenesis.
My analysis data concluded that the FOX family of transcription factors developed therapeutics and preventives strategy in mammals. In contrast, the restricted expression of FOXP3 in the cell is an immune-privilege and therapeutic strategy. The ultimate transcriptional regulation of FOXP3 in tumour cells may represent a novel mechanism in the immune system.
The author is grateful to the editor and anonymous reviewers for valuable comments and suggestions. Also thanks to the Assam University, Silchar, Assam, India for providing the requisite lab facilities in carrying out this research work.
Citation: Choudhury S (2020) In Silico Analysis of the FOXP3 Transcription Factor Associated with T-Cell Oncogenesis. J Cancer Res Immunooncol.6:120. DOI: 10.35248/2684-1266.20.6.120
Received: 10-Jul-2020 Accepted: 24-Jul-2020 Published: 31-Jul-2020 , DOI: 10.35248/2684-1266.20.6.120
Copyright: © 2020 Choudhury S. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.