Journal of Information Technology & Software Engineering
Open Access
ISSN: 2165- 7866
ISSN: 2165- 7866
Research - (2022)Volume 12, Issue 3
In brain tumors treatment planning and quantitative evaluation, determining the tumors extent is a major challenge. Noninvasive Magnetic Resonance Imaging (MRI) has developed as a front-line diagnostic technique for brain malignancies without the use of ionizing radiation. Segmenting the extent of a brain tumor manually from 3D MRI volumes is a time-consuming process that relies greatly on operator competence. For correct tumors extent evaluation, a reliable fully automated brain tumors segmentation approach is required in this scenario. We present a completely automated brain tumors segmentation method based on U-Net deep convolutional networks in this paper. The Multimodal Brain Tumor Image Segmentation (BRATS 2015) datasets were utilized to test our approach, which included 220 high-grade brain tumors.
Magnetic resonance imaging; Deep learning; U-Net deep; 3D MRI
The brain, which contains billions of cells, is one of the most complex organs in the human body. When cells divide uncontrollably, they form an abnormal group of cells around or inside the brain, which is known as a brain tumor. Benign tumors are non-cancerous and therefore considered less aggressive. They start in the brain and grow slowly, and they cannot spread to other parts of the body [1]. Malignant tumors, on the other hand, are cancerous and have no defined boundaries. In both the detection and treatment phases, researchers have relied on brain Magnetic Resonance Imaging (MRI) as one of the best imaging techniques for detecting brain tumors and modeling tumors progression. Because of the high resolution of MRI images, they have a significant impact on automatic medical image analysis. Using brain MRI images as data sets and the CNN algorithm to extract and test data, we will apply the deep learning concept to perform automated brain tumors detection [2,3].
The segmented brain tumors extent can eliminate confounding structures from other brain tissues and thus provide a more accurate classification for the sub-types of brain tumors and inform the subsequent diagnosis; the accurate delineation is crucial in radiotherapy or surgical planning, from which not only the brain tumor extend but also surrounding healthy tissue has been outlined. Segmentation is still done manually by human operators in current clinical practice [4]. Manual segmentation is a time-consuming task that typically involves slice-by-slice procedures, and the results are highly reliant on the operators' knowledge and subjective decision-making. Furthermore, even when using the same operator, consistent results are difficult to achieve. A fully automatic, objective, and repeatable segmentation method is highly desired for a multimodal, multi-institutional, and longitudinal clinical trial [5].
This project's main goal is to create a cutting-edge Convolutional Neural Network (CNN) model for classifying skin lesion images into cancer types. On the dataset made available by the International Skin Imaging Collaboration, the model is trained and tested (ISIC). The model can be used to examine a lesion image and determine whether or not it is dangerous at an early stage.
For image and audio applications, such as [6-8], etc., unsupervised feature learning methods and deep learning have been widely used. These techniques have shown a lot of promise in these domains for automatically representing the feature space using unlabeled data to improve the accuracy of subsequent classification tasks. These capabilities have been extended to allow learning in very high dimensional feature spaces using additional data properties. A method to scale unsupervised feature learning and deep learning methods to high dimensional and full sized images by using image characteristics such as locality and stationary of images. Similarly, an unsupervised feature learning method (specifically Reconstruction Independent Subspace Analysis) to classify histological image signatures and tumor architecture in the context of cancer detection [9]. Unsupervised feature learning methods have not been applied to gene expression analysis to our knowledge (it should be noted that Le's method is still used for images, not gene expression). The extremely high dimensionality of gene expression data, the lack of sufficient data samples, and the lack of global known characteristics such as locality in gene expression data are some of the reasons for this, which limit the applicability of techniques such as convolution or pooling, which have been highly successful in the above-mentioned image data applications [10,11].
We try to solve the dimensionality problem in gene expression data with the method proposed here. In our method, we first use PCA to reduce the dimensionality of the feature space, and then apply the result of PCA as a compressed feature representation that still encodes the data available in the sample set, as well as some randomly selected original gene expressions (i.e. original raw features) as a more compact feature space to either a one or a multi-layered sparse auto-encoder to find a sparse representation for data that will be used for classification (Figure 1) depicts the overall process of developing and training a system to detect and classify cancer based on gene expression data [12-15]. The approach proposed here, as shown in the figure, is divided into two parts: feature learning and classifier learning.
A file containing one or more records is referred to as a data set. A programed operating on z/OS uses records as the basic unit of information [16].
A data set is a named collection of records. Data sets can contain information such as medical records or insurance records for use by a system application. Data sets are also used to store information that is required by applications or the operating system, such as source programmers, macro libraries, and system variables or parameters [17,18]. You can print or show data sets that contain readable text on a console if they contain readable text (many data sets contain load modules or other binary data that is not really printable). Cataloging data sets is possible.
In the last few decades, medical image analysis and classification has become a burgeoning field, with health-related applications that could aid medical experts and radiologist in diagnosing chronic diseases like chest cancer, brain tumors, diabetes prediction, and cardiac murmur evaluation [19]. A hybrid approach based on k-means clustering and a fine-tuned CNN model with synthetic data augmentation is presented in this paper. The proposed technique consists of three main stages: (a) normalizing and unbiased the intensities of MR modalities; (b) segmenting tumor area using k-means clustering; and (c) synthetic data augmentation followed by deep features extraction from ROI and classification of brain tumor into relevant benign or malignant categories using a finetuned CNN model. Convolution, ReLU Layer, pooling, flattening, and full connection were all used in the CNN model's training [20-23].
Dataset Brain MRI images of 153 patients, including normal and brain tumor patients, were used in this study. The images collected included brain images of 80 healthy patients after the doctor's examination and diagnosis. There are 1321 images total, with 56 for testing and 515 for train data. 73 patient tumors include 571 images, with 170 test images and 1151 train images. Patients with brain tumors account for a small percentage of the total number of patients.
86 women and 68 men, ranging in age from 8 to 66 years old, were diagnosed with the disease. 1892 images were collected from a total of 153 patients, 1666 for train data and 226 for test data. The original size of the collected images was 512512 pixels.
Some areas of fat in photographs are mistakenly identified as tumors, or tumors may not be visible to the physician; the most precise diagnosis is entirely dependent on the physician's skill. The CNN was used to detect tumors using brain images in this paper. The images gathered from imaging centers had additional margins. To keep the images from becoming noisy, these margins were cropped. One of the primary motivations for combining the feature extraction technique with the CNN is to retrieve image feature extraction in order to improve the network's accuracy [24]. According to the results of the CNN on the initial images, a new method combining the Clustering algorithm for feature extraction and CNN is proposed in this study to improve the network accuracy.
Method of obtaining features (C) A clustering method called central clustering. This algorithm has a duplicate procedure that iteratively tries to get points as cluster centers, which are actually the same mean points belonging to each cluster, for a fixed number of clusters. Assign each sample data set to a cluster with the shortest distance between the data and the cluster's center. The cluster centers are first chosen at random in the basic version of this method [25,26]. According to the degree of similarity, the points are assigned to the cluster centers, resulting in new clusters. The first-order clustering algorithm is used to extract features from the data in this paper. The image produced by applying the clustering algorithm to the image is shown in Figures 1 and 2.
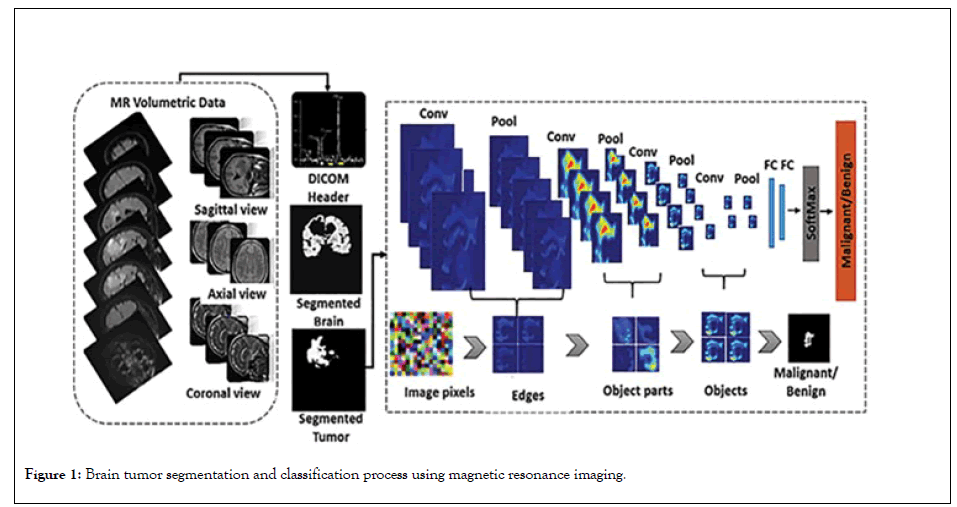
Figure 1: Brain tumor segmentation and classification process using magnetic resonance imaging.
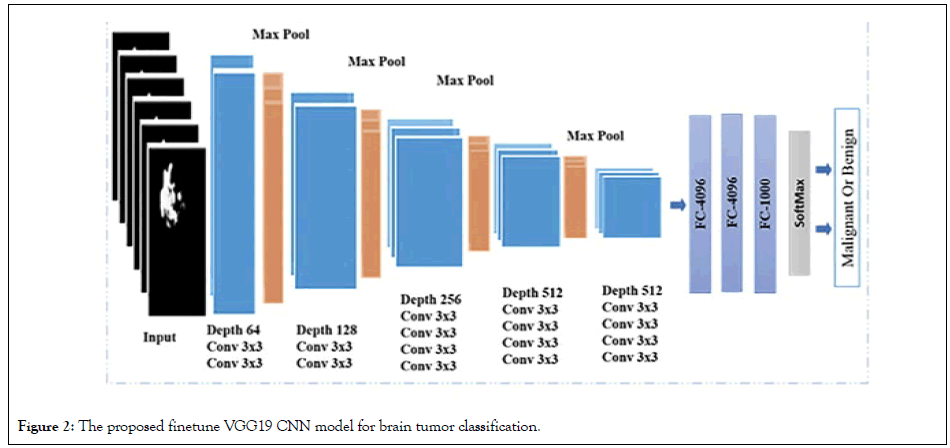
Figure 2: The proposed finetune VGG19 CNN model for brain tumor classification.
D. Neural network convolutional initially, no feature extraction methods were used when the images were fed into the CNN. The input images are 227 227 pixels wide at first. To identify and classify the images, the Alex net architect was used, which consisted of 5 Convolutional layers and 3 layers of Sub-sampling layers, Normalization layers, Normalization layers, Fully Connected layers, and the classification layer [21]. 4096 neurons are present in the fully connected layers. This layer has two classes: brain tumor patients and healthy people
Deep learning methods were used instead of traditional machine learning in a large portion of the recently published brain tumour classification literature. For the classification of brain tumour tasks, many researchers used pre-trained CNN architectures that were fine-tuned. Abrasion-based architectures have been proposed/ tested and trained by some data scientists. The majority of the CNN architectures were 2D, but some were 3D. The main issue in the age of deep learning is the lack of large enough labeled MRI data for CNN training. Synthetic data augmentation was used to solve this problem. For brain tumour classification, proposed three convolutional auto encoders (for three MR sequences), fusion, and finally a fully linked layer. After extensive data augmentation, Sajjad, et al. fed the tumor area to a pre-trained VGG-19 CNN, which was fine-tuned for grade classification [6]. Rehman, et al. [4] proposed combining pre-trained CNN models (VGG16 VGG19) to classify multimodal automated brain tumors using linear contrast stretching, a transition of learning-based extraction functions, and correntropy-based collection [4]. Finally, to detect a brain tumor, the fused matrix was fed to an extreme learning machine (ELM). For BraTs 2015, BraTs 2017, and BraTs 2018, the proposed methodology was validated on the BraTS data sets, with precision of 97.8, 96.9, and 92.5%, respectively (Figures 3 and 4). Figure 3 depicts the model effect after the end of a successful run, whereas Figure 4 depicts the model effect throughout.
Figure 3: Data set of infected brain.
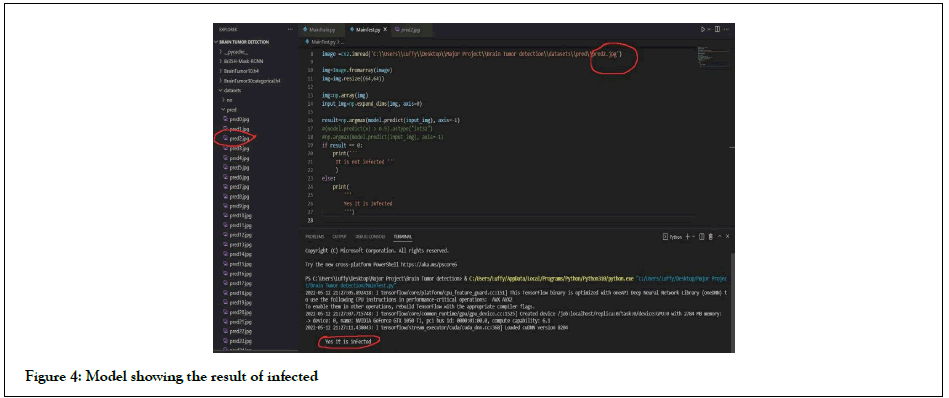
Figure 4: Model showing the result of infected
Various types of features were extracted and trained using various machine learning techniques to improve the prediction accuracy of overall survival for glioma patients in MRI. Deep feature extraction using pre-trained Alex Net and Linear Discriminant yielded the highest classification accuracy. Despite the fact that texture features are widely used in tumor classification, their accuracy is only 46%. For two classification classes, histogram features had a precision of 68.5 percent; this low precision could be due to noise. The number of coils in the imaging system determined the noise model for MRI images. In a single-coil system, the noise is represented by a Rician distribution, while in a multi-coil (parallel) imaging system, it is represented by a non-central chi distribution. The data provided was noisy, as shown in Figures 5 and 6. The single-coil system assumes that an MRI image's real and imaginary parts are uncorrelated Gaussian distributions with zero mean and equal variance. The Rician distribution is, in fact, a subset of the noncentral chi distribution [20-22].
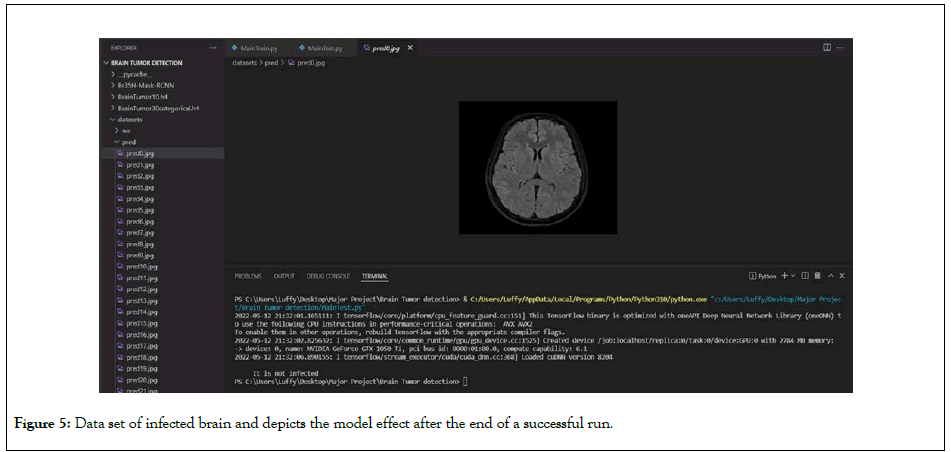
Figure 5: Data set of infected brain and depicts the model effect after the end of a successful run.
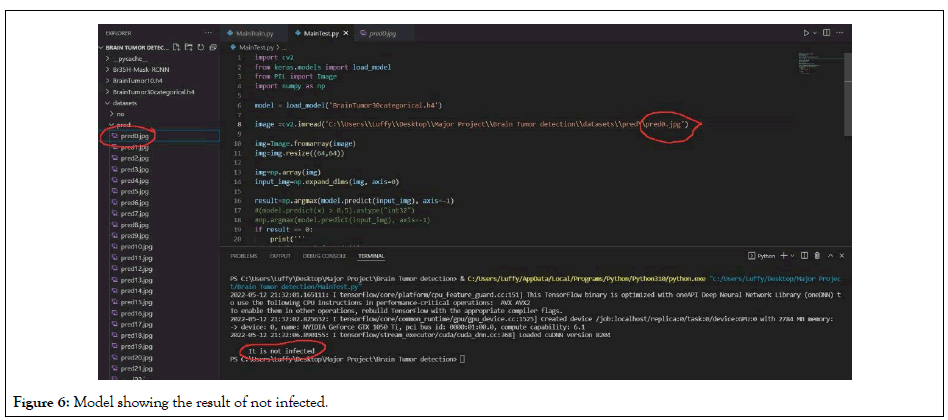
Figure 6: Model showing the result of not infected.
DE noising the data and testing the histogram features, as well as other types of proposed features, is one recommendation for future research. In addition, texture features for each sample were extracted using specific slices that contained the three tumor regions. To improve deep feature classification accuracy, the specific patch size that contains the tumor could be used only to extract the 2D and 3D deep features, based on classification for three classes; then classification for two classes could be tested as well.
[CrossRef] [Google Scholar] [Pubmed]
[CrossRef] [Google Scholar] [Pubmed]
[CrossRef] [Google Scholar] [Pubmed]
[CrossRef] [Google Scholar] [Pubmed]
[CrossRef] [Google Scholar] (All versions) [Pubmed]
[CrossRef] [Google Scholar] [Pubmed]
[CrossRef] [Google Scholar] [Pubmed]
[CrossRef] [Google Scholar] (All versions) [Pubmed]
[CrossRef] [Google Scholar] (All versions)
[CrossRef] [Google Scholar] (All versions)
[CrossRef] [Google Scholar] [Pubmed]
Citation: Konwar P, Bhadra J, Dutta MJ, Dowari J (2022) Detecting Brain Tumour in Early Stage Using Deep Learning. J Inform Tech Softw Eng. 12:297.
Received: 24-Mar-2022, Manuscript No. JITSE-22-17474; Editor assigned: 28-Mar-2022, Pre QC No. JITSE-22-17474 (PQ); Reviewed: 15-Apr-2022, QC No. JITSE-22-17474; Revised: 22-Apr-2022, Manuscript No. JITSE-22-17474 (R); Published: 29-Apr-2022 , DOI: 10.35248/2329-9096-22.12.297
Copyright: © 2022 Konwar P, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.