Immunological Disorders and Immunotherapy
Open Access
ISSN: 2593-8509
ISSN: 2593-8509
Research Article - (2023)Volume 8, Issue 1
Objective: Curcuma has long been used as a Chinese herbal medicine for the treatment of chronic liver disease, and curcumol is its main active ingredient. This study explores the molecular mechanism of curcumol regulating glutamine metabolism, hoping to provide new ideas for the prevention and treatment of chronic liver disease.
Methods: We used LPS to activate hepatic stellate cells. The effects of curcumol on glutamine metabolism and cell proliferation were detected by kit. The effects of curcumol on GLS1, senescence-related proteins and extracellular matrix expression were observed by Western blotting and RT-PCR. The effect of curcumol on cell cycle was detected by flow cytometry. Furthermore, after overexpression of GLS1, we observed the effects of curcumol on glutamine metabolism, expression of related molecules and cell cycle.
Results: Curcumol significantly inhibited cell proliferation, glutamine metabolism, GLS1 and extracellular matrix expression, and significantly promoted cell senescence. When we further over express GLS1, the inhibitory effect of curcumol on glutamine metabolism and the effect of promoting cell senescence were also saved.
Conclusions: Curcumol targets GLS1, inhibits glutamine metabolism and induces senescence of hepatic stellate cells, which may be one of the important mechanisms of its anti-hepatic fibrosis. GLS1 may be an important target site for the prevention and treatment of chronic liver disease.
Curcumol; Hepatic fibrosis; Glutamine; GLS1; Hepatic stellate cells
Hepatic fibrosis is the response of various chronic liver injury factors to the liver, and further development will lead to irreversible changes in liver structure and function [1]. The main pathological feature of hepatic fibrosis is the massive deposition of Extra Cellular Matrix (ECM) [2]. Hepatic Stellate Cells (HSC) activation is an important event in liver fibrosis and a major source of ECM [3]. Therefore, targeting HSC is still a hot issue in the prevention and treatment of liver fibrosis. There is a huge energy demand for HSC activation, leading to a new adaptation of HSC metabolism [4]. Energy needs are met by protein metabolism in addition to sugar and fat metabolism reprogramming. Some studies have found that by comparing the differentially expressed metabolic genes between quiescent HSC and activated HSC, it is found that only 6% of genes are involved in carbohydrate metabolism, while 38% are related to protein metabolism, among which the difference in glutamine metabolism is the most significant [5]. It is suggested that protein metabolism may be more important than glucose metabolism in the process of cell proliferation and activation. Glutamine is the most abundant amino acid in the blood and has been widely reported as the second carbon source after glucose in energy production and anabolism [6]. Glutamine metabolism mainly refers to deamination under the catalysis of Glutaminase (GLS) to generate glutamate, which is then converted into α-Ketoglutarate (α-KG) and enters the tricarboxylic acid cycle for metabolic energy production [7]. Therefore, targeting glutamine metabolism may be a new strategy to inhibit HSC activation.
Curcuma (Curcuma kwangsiensis S.G. Leet and C. F. Liang) is the rhizome of the ginger plant Curcuma or Turmeric. It is used as a medicinal plant material in Guangxi, China. It is called “Ginghgvun” by the local folk and is included in the 2020 edition of the “Chinese Pharmacopoeia. [8]” It was first recorded in “The Theory of Medical Properties.” In traditional Chinese medicine it is employed in promoting qi and blood, eliminating accumulation of toxins, and relieving pain. It has been used for the treatment of chronic liver disease for many years [9].
The research group has previously confirmed that curcumol can inhibit liver inflammation and improve liver microcirculation to play an exact anti-hepatic fibrosis role [10,11]. Recent studies have found that curcumol's anti-hepatic fibrosis effect is closely related to its regulation of glutathione metabolism [12]. Because glutamine is an upstream substance of glutathione, it indicates that curcumol may be closely related to glutamine metabolism. However, the role of glutamine metabolism in the prevention and treatment of liver fibrosis has not been fully elucidated. Therefore, this study hoped to explore the molecular mechanism of curcumol's anti-hepatic fibrosis effect based on glutamine metabolism.
Reagents and antibodies
Aladdin Chemical Co., Ltd. provided the curcumol (Shanghai, China). Abcam provided the following primary antibodies against GLS1, P16, P21 and Hmg1 (Cambridge, UK). Cell Signaling Technology sold antibodies against Cyclin D1, Cyclin E1, CDK4, α-SMA, and collagen I (Denver, MA, USA). HyClone provided Dulbecco's Modified Eagle's Medium (DMEM) and Fetal Bovine Serum (FBS) (Grand Island, NY, USA).
Cell culture and treatment
The HSC was acquired from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). The cells were cultured in standard DMEM-basic, supplemented with 10% calf serum (from Gibco, Grand Island, NY, USA) and maintained at 37°C in a humidified incubator of 5% CO2 and 95% air. HSCs were cultured until they adhered well, and when the cell density was 90%, after digestion with 0.25% trypsin, the cells were divided into blank control group, LPS group (1ug/mL) [13], curcumol 12.5 mg/L group, curcumol 25 mg/L group, curcumol 50 mg/L group [14]. Except for the blank control group, each group was first treated with LPS for 24 h, and then treated with corresponding drugs for 48 h. The blank control group was cultured with culture medium throughout [15].
Cell viability assay
To evaluate cell viability, the CCK8 Cell Counting Kit (ab228554, Abcam) was utilized. HSC cells were seeded in a 96-well plate (CLS3300, Corning) and treated with the cytotoxic chemicals for the timeframes specified. The 10 μl CCK8 reagents were applied to each well and incubated for 4 hours at 3°C in 5% CO2 before being measured at 450 nm using the Tecan Safire2 Multi-detection Microplate Reader (Morrisville, NC).
Measurement of levels of glutamine, glutamic acid, ATP, GSH and α-KG
Glutamine and glutamate concentrations were determined using glutamine/glutamate assay kits (GLN1-1KT, Sigma). Relative glutathione levels were measured using GSH assaykit (Cayman, Ann arbor, MI, USA) following the manufacturer's instruction. The relative content of ATP and α-KG was detected using abcam kit (Cambridge, UK).
Flow cytometry
HSCs were transferred to 6-well plates overnight and processed accordingly. After 48 h, the cells were washed two to three times with Phosphate Buffered Saline (PBS), before being digested using 0.25% trypsin, washed twice again with PBS and transferred into eppendorf tubes. The cells were counted and 1×106 cells were resuspended with 100 μl PBS before 500 μl cooled 75% ethanol was added after centrifugation. Cell cycle distribution was detected using the CytoFLEX Flow Cytometer (Beckman Coulter, Inc.) and the results were analysed using ModFitLT 5 software (Verity Software House, Inc.).
Reverse Transcription-quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted using total RNA extraction reagent (Sigma, St. Louis, MO, USA), and primers were created using Primer 5.0 software by Wuhan Bafeier Biotechnology Company. cDNA was generated using a reverse transcription kit (Promega, USA). The reaction system was set up as follows: 95°C for 1 minute; 95°C for 10 seconds, 60°C for 30 seconds, 40 cycles, using β-actin as the internal reference, and the PCR products were collected for gel imaging analysis. Each sample's relative mRNA expression level = 2-△△Ct. The primers are shown in Table 1.
| Gene | Forward | Reverse |
|---|---|---|
| CollagenⅠ | 5′-CCAGTGGCGGTTATGACTTC-3′ | 5′-GCTGCGGATGTTCTCAATCT-3′ |
| α-SMA | 5′-CTGCTACTGGTTGTTCTTGTGGC-3′ | 5′-GTTCTGTGCCTCTCCATTCTTAG-3′ |
| GLS1 | 5′-GCAACAGCGAGGGCAAAGA-3′ | 5′-TTCAACCTGGGATCAGACG-3′ |
| P16 | 5′-GGGTCGGGTAGAGGAGGTG-3′ | 5′-ACCGTAACTATTCGGTGCGT-3′ |
| P21 | 5′-CACCACTGGAGGGTGACTTC-3′ | 5′-ATCTGTCATGCTGGTCTGCC-3′ |
| Hmgα1 | 5′-GGAAAAGGACGGCACTGAGA-3′ | 5′-GGCTCCTGACTCCCTACCA-3′ |
| CyclinD1 | 5′-CGGACTACAGGGGAGTTTTG-3′ | 5′-AGGAGGTTGGCATCGGGGT-3′ |
| CyclinE1 | 5′-AGTACCCACAGCAGGTCTTC-3′ | 5′-CTGCATCAACTCCAACGAGG-3′ |
| CDK4 | 5′-GCCAGTCTGATTTGTGGGCC-3′ | 5′-GCCAGTTGTTTTTCTGCCAC-3′ |
| β-actin | 5′-TGTCCCTGTACACCTCTG-3′ | 5′-ATGTCACGCACGATTTCCC-3′ |
Table 1: Primer sequence list.
Western blot analysis
Cells were harvested by centrifugation at 12,000 rpm for 15 min at 4°C and lysed in RIPA buffer (Sigma-Aldrich). Protein concentrations were determined using a Bicinchoninic Acid (BCA) protein assay kit (Thermo Scientific, CA, USA). After boiling for 10 min in loading buffer, the samples (20 μg protein per lane) were separated by 10% or 12% SDS-PAGE. The proteins were transferred to nitrocellulose membranes with 0.45- μm diameter pores. The membranes were blocked with 5% nonfat dry milk for 1h at room temperature and then incubated with primary antibodies, including P16(1:1000), P21(1:1000), Hmgα1(1:1000), Cyclin D1(1:1000), Cyclin E1(1:1000), CDK4(1:1000), α-SMA (1:1000), collagen I (1:1000), and GLS1 (1:1000) antibodies. After incubation overnight, the membranes were washed 4 times in 1X Tris-Buffered Saline, 0.1% Tween 20 Detergent (TBST) and incubated with secondary antibody (1:1000) for another 1 h at room temperature. The protein bands were visualized with Immobilon Western HRP Substrate (Millipore, MA, USA). Densitometric analyses of the bands were performed using ImageJ software.
Statistical analysis
The statistical analysis was performed with IBM SPSS Statistics 25 (IBM, NY, USA). Data are expressed as the means ± Standard Deviation (SD). Groups were compared with one-way or two-way Analysis of Variance (ANOVA), followed by Bonferroni post hoc analysis. A value of P<0.05 was considered to indicate statistical significance.
Curcumol inhibits the proliferation of HSC and reduces the expression of ECM
The results of the CCK8 assay showed that curcumol significantly inhibited the proliferation of hepatic stellate cells, as detailed in Figure 1A. RT-PCR examination revealed that curcumol significantly inhibited α-SMA and CollagenⅠmRNA expression, as detailed in Figure 1B. Immunoblotting assays revealed that curcumol significantly inhibited α-SMA and CollagenⅠprotein expression, as detailed in Figure 1C. In summary, the curcumol has an inhibitory effect on the activation of hepatic stellate cells.
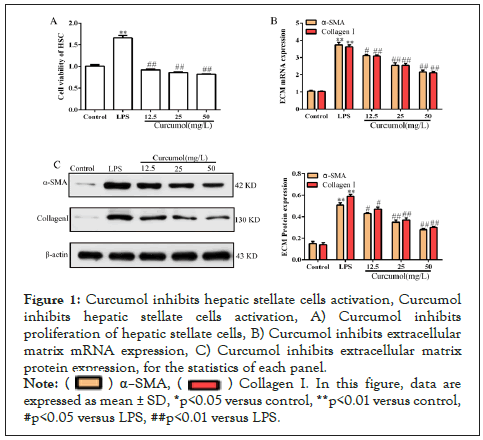
Figure 1: Curcumol inhibits hepatic stellate cells activation, Curcumol inhibits hepatic stellate cells activation, A) Curcumol inhibits proliferation of hepatic stellate cells, B) Curcumol inhibits extracellular matrix mRNA expression, C) Curcumol inhibits extracellular matrix protein expression, for the statistics of each panel.
Note: ( ) α–SMA, (
) α–SMA, ( ) Collagen I. In this figure, data are expressed as mean ± SD, *p<0.05 versus control, **p<0.01 versus control, #p<0.05 versus LPS, ##p<0.01 versus LPS
) Collagen I. In this figure, data are expressed as mean ± SD, *p<0.05 versus control, **p<0.01 versus control, #p<0.05 versus LPS, ##p<0.01 versus LPS
Curcumol inhibits GLS1 expression and glutamine metabolism
During the activation of hepatic stellate cells, more energy and substrates are required and glutamine metabolism can provide sufficient energy and biosynthesis, as detailed in Figure 2A.
Curcumol significantly decreased the level of intracellular ATP, as detailed in Figure 2B. In addition, curcumol decreased glutamate levels and increased glutamine levels, as shown in Figure 2C-2D. The above results suggest that curcumol blocks the conversion of glutamine to glutamate. The key enzyme for the conversion of glutamine to glutamate is GLS1, and molecular biology experiments have found that curcumol can significantly inhibit its expression, as detailed in Figure 2E. Glutamate can be further metabolized into α-KG and GSH to maintain cell energy metabolism and homeostasis. At the same time, curcumol could reduce the expression of α-KG and GSH in hepatic stellate cells, as shown in Figure 2F-2G. The above results showed that glutamine metabolism in hepatic stellate cells was blocked by curcumol.
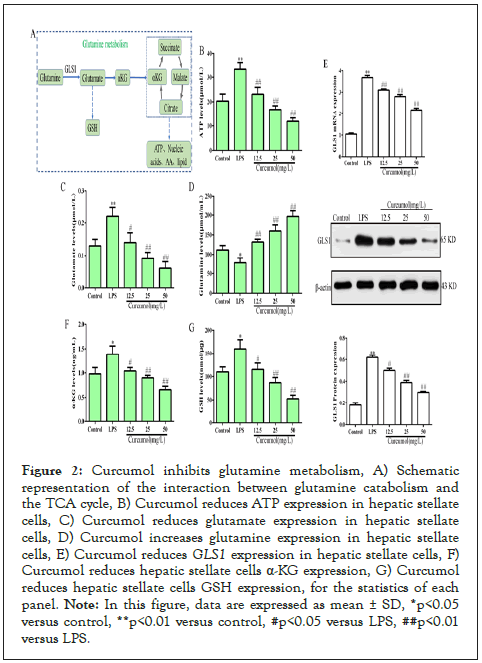
Figure 2: Curcumol inhibits glutamine metabolism, A) Schematic representation of the interaction between glutamine catabolism and the TCA cycle, B) Curcumol reduces ATP expression in hepatic stellate cells, C) Curcumol reduces glutamate expression in hepatic stellate cells, D) Curcumol increases glutamine expression in hepatic stellate cells, E) Curcumol reduces GLS1 expression in hepatic stellate cells, F) Curcumol reduces hepatic stellate cells α-KG expression, G) Curcumol reduces hepatic stellate cells GSH expression, for the statistics of each panel.
Note: In this figure, data are expressed as mean ± SD, *p<0.05 versus control, **p<0.01 versus control, #p<0.05 versus LPS, ##p<0.01 versus LPS.
Curcumol induces senescence of HSC
The results of molecular biology experiments showed that curcumol significantly increased the expression of aging marker molecule (P16, P21, Hmgα1), as detailed in Figure 3A. At the same time, flow cytometry showed that curcumol could block the cell cycle more in G0/G1 phase, as detailed in Figure 3B. And curcumol can reduce the expression of G1-related regulatory factors (CyclinD1, CDK4, CyclinE1), as detailed in Figure 3C. These results suggest that curcumol can induce senescence of hepatic stellate cells.
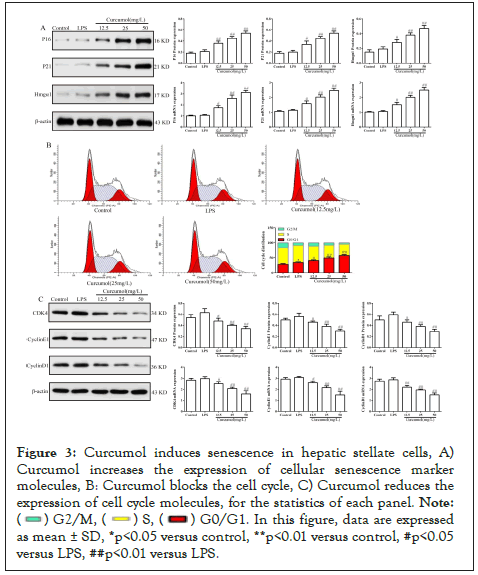
Figure 3: Curcumol induces senescence in hepatic stellate cells, A) Curcumol increases the expression of cellular senescence marker molecules, B: Curcumol blocks the cell cycle, C) Curcumol reduces the expression of cell cycle molecules, for the statistics of each panel.
Note: ( ) G2/M, (
) G2/M, ( ) S, (
) S, ( ) G0/G1. In this figure, data are expressed as mean ± SD, *p<0.05 versus control, **p<0.01 versus control, #p<0.05 versus LPS, ##p<0.01 versus LPS.
) G0/G1. In this figure, data are expressed as mean ± SD, *p<0.05 versus control, **p<0.01 versus control, #p<0.05 versus LPS, ##p<0.01 versus LPS.
Curcumol inhibited glutamine metabolism was saved after GLS1 was overexpressed
Curcumol significantly reduced ATP content and glutamate content in hepatic stellate cells, but this effect was rescued by overexpression of GLS1, see Figure 4A-4B for details. Curcumol increased glutamine content, but overexpression of GLS1 reversed the increase in glutamine, see Figure 4C for details. Overexpression of GLS1 increased both α -KG and GSH content, reversing the effect of curcumol on α-KG and GSH down-regulation effect of curcumol, as detailed in Figure 4D-4E. Based on the above results, the results suggest that GLS1 is a key target in the regulation of glutamine metabolism by curcumol.
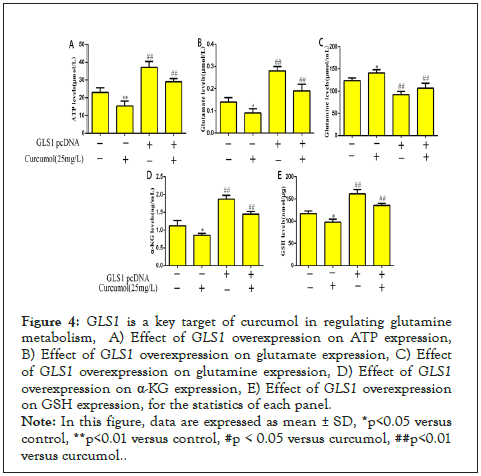
Figure 4: GLS1 is a key target of curcumol in regulating glutamine metabolism, A) Effect of GLS1 overexpression on ATP expression, B) Effect of GLS1 overexpression on glutamate expression, C) Effect of GLS1 overexpression on glutamine expression, D) Effect of GLS1 overexpression on α-KG expression, E) Effect of GLS1 overexpression on GSH expression, for the statistics of each panel.
Note: In this figure, data are expressed as mean ± SD, *p<0.05 versus control, **p<0.01 versus control, #p < 0.05 versus curcumol, ##p<0.01 versus curcumol.
Curcumol induced cell senescence was saved after GLS1 was overexpressed
Curcumol increased the expression of P16, P21 and Hmga1 in hepatic stellate cells, but this effect was saved by overexpression of GLS1, as detailed in Figure 5A. Curcumol decreased the expression of CyclinD1, CDK4 and CyclinE1, but reversed the increase of cyclin after overexpression of GLS1, as detailed in Figure 5B. Curcumol blocks hepatic stellate cells in G0/G1 phase. The overexpression of GLS1 reversed the cell cycle arrest effect of curcumol, as detailed in Figure 5C. Based on the above results, curcumol targeting GLS1 is the key target to induce the senescence of hepatic stellate cells.
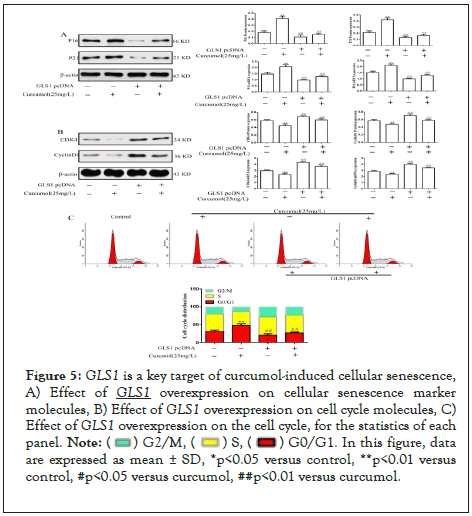
Figure 5: GLS1 is a key target of curcumol-induced cellular senescence, A) Effect of GLS1 overexpression on cellular senescence marker molecules, B) Effect of GLS1 overexpression on cell cycle molecules, C) Effect of GLS1 overexpression on the cell cycle, for the statistics of each panel.
Note: ( ) G2/M, (
) G2/M, ( ) S, (
) S, ( ) G0/G1. In this figure, data are expressed as mean ± SD, *p<0.05 versus control, **p<0.01 versus control, #p<0.05 versus curcumol, ##p<0.01 versus curcumol.
) G0/G1. In this figure, data are expressed as mean ± SD, *p<0.05 versus control, **p<0.01 versus control, #p<0.05 versus curcumol, ##p<0.01 versus curcumol.
Various chronic liver injury factors act on the liver, which makes HSC activate and transform into Myofibroblasts (MFB). MFB secretes a large amount of collagen and synthesizes, which leads to liver fibrosis [16]. Myofibroblasts derived from activated hepatic stellate cells are the ECM major producers of the fibrous matrix that accumulates with cirrhosis, thus limiting the accumulation of Myofibroblasts in the damaged liver may prevent cirrhosis [17]. Theoretically, this could be achieved by inhibiting activation of HSC, promoting the return of activated HSC to a quiescent phenotype, or by killing activated HSC, thereby reducing MFB production [18]. The mechanisms that naturally orchestrate these changes in HSC fate are poorly understood but would seem to be ideal therapeutic targets to limit accumulation of MFB and prevent excessively fibrogenic repair. Metabolic pathways with increased bioenergetic and biosynthetic demands upon HSC activation are attractive therapeutic options for the prevention of liver fibrosis.
Earlier studies found that the activation mechanism of HSC is quite complex. On the one hand, damaged epithelial cell paracrine signals, metabolic disorders, and hepatitis virus products can directly or indirectly induce HSC activation [19-21]. TGF-β/Smad and PDGF-pR/ERK are the two main signaling pathways that promote HSC activation. On the other hand, studies reported that after activation of hepatic stellate cells, their bioenergetic and biosynthetic requirements increased dramatically, similar to those of highly proliferative cancer cells [4,22]. Glutaminolysis is an important metabolic feature other than Warburg during HSC activation [23]. Glutamine plays a wide range of roles in cellular metabolism and is involved in Tricarboxylic Acid (TCA) cycle replenishment and in the biosynthesis of nucleotides, Glutathione (GSH), and other non-essential amino acids [24]. Since Glutaminase 1 (GLS1), a key mitochondrial enzyme that catalyzes the deamidation of glutamine, was first discovered in the kidney in 1958 [25]. Glutamine is decomposed into glutamate under the action of GLS1, leading to the synthesis of glutathione, which plays an important role in cellular redox homeostasis [26]. Some studies have found that cellular Myelocytomatosis oncogene (c-Myc) further promotes glutamine metabolism by inhibiting the expression of miR-23a, thereby enhancing the expression of mitochondrial glutaminase [27]. Studies have found that in a mouse model of liver fibrosis caused by Non-Alcoholic Steatohepatitis (NASH), with the occurrence of liver fibrosis, the increased level of glutamine breakdown is synchronized with the severity of liver fibrosis [23].The Hedgehog pathway is involved in the activation of HSC and initiates the expression of fibrosis-related genes through the downstream effector Gli. The Hedgehog pathway is also involved in the breakdown of glutamine, and a decrease in the expression levels of glutaminase and peroxidase was observed after inhibition of this pathway [28]. To sum up, the regulation of glutamine metabolism is an effective strategy for the prevention and treatment of liver fibrosis.
Cellular senescence refers to a relatively stable state of programmed cellular response following the irreversible deregulation of the cell cycle and loss of proliferative capacity by certain specific stimuli [29]. Cellular senescence can act as an inherent cytoprotective mechanism to block the proliferation of damaged or cancerous cells [30]. Of interest, regardless of the specific stimuli that induce cellular senescence, the majority of cellular senescence is regulated by the p53/P21 signalling pathway and the P16/PRB signalling pathway [31]. External stimulation of senescence signal can activate p53 and induce the stable expression of its target gene P21, which is a classical cell cycle inhibitor, which can specifically bind to cyclin- dependent kinase in G1 phase of cell cycle. Finally, the cell cycle is stagnated in G1 phase, which induces cell senescence [32]. P16 binds to CDK4/6 and inhibits its kinase activity, which in turn blocks pRb phosphorylation, leaving pRb in an inhibited state of activity, resulting in the inability to express many key cell cycle target genes and thus causing cell cycle arrest [33]. It has been shown that rhodopsin induces hepatic stellate cell senescence and reduces liver fibrosis via the nuclear receptor Nur77 [34]. To sum up, inducing senescence of hepatic stellate cells is a new way to reverse hepatic fibrosis.
In this study, we first used CCK8 to detect the effect of curcumol on the proliferation of hepatic stellate cells, and found that curcumol could inhibit the proliferation of hepatic stellate cells. Further molecular biological detection showed that curcumol could inhibit the expression of extracellular matrix, indicating that curcumol could inhibit the activation of hepatic stellate cells. Curcumol could inhibit glutamine metabolism and molecular biology test showed that curcumol could block cell cycle and induce cell senescence. Then we overexpressed GLS1 and found that curcumol inhibited glutamine metabolism and induced cell senescence was saved, indicating that GLS1 is the key target of curcumol to regulate glutamine metabolism. In general, curcumol inhibits glutamine metabolism in hepatic stellate cells by targeting GLS1, induces cell senescence and reduces the expression of extracellular matrix, which may be its mechanism of anti-hepatic fibrosis. However, there are some problems in this study, and there is no further exploration of the relationship between glutamine metabolism and cell senescence, nor the regulatory mechanism of the upstream of GLS1. Next, our team will explore whether glutamine induces cell senescence through energy metabolism or oxidative stress, and will further clarify the epigenetic regulation mechanism in the upstream of GLS1.
Curcumol inhibits glutamine metabolism by targeting GLS1, thereby inhibiting the activation of hepatic stellate cells and reducing the production of extracellular matrix, exerting an anti- fibrotic effect. In summary, GLS1 can be considered as a potential candidate target for drug treatment of liver fibrosis. However, more work is needed to be undertaken to clarify the definite mechanism on YAP regulation of NCOA4 by curcumol. This study is helpful for curcumol to become a novel effective natural compound for the prevention and improvement of NAFLD.
All authors have carefully read and revised the full text and agreed to its publication.
The data used and analyzed in this study are included within the article.
This study was funded by the National Natural Science Foundation of China (Grant Nos. 82204755, 81960751, and 81660705), the Guangxi Young and Middle-aged Teachers’ Research Ability Improvement Project (Grant No. 2022KY1667), the Guangxi Zhuangyao Pharmaceutical Key Laboratory (Grant Nos. GXZYZZ2019-1, GXZYZZ2020-07), the Guangxi Natural Science Foundation Youth Project (Grant No. 2020GXNSFBA297094), and the Guangxi University of Traditional Chinese Medicine School-level Project Youth Fund (Grant No. 2022QN008). Faculty of Chinese Medicine Science Guangxi University of Chinese Medicine Research Project(2022MS008, 2022QJ001)
The authors declare that there are no financial conflicts of interest.
Xuelin Duan and Tiejian Zhao contributed equally to this work. ot applicable.
We thank the Guangxi University of Chinese Medicine (Guangxi, China) for providing laboratory equipment and technological support.
Mechanism of action of curcumol targeting GLS1 to inhibit glutamine metabolism to induce cellular senescence to prevent liver fibrosis.f China (Grant number 81802777).
Citation: Duan X, Zhao T, Wang J, Wang J, Zheng Y (2023) Curcumol Targets Gls1 to Regulate Glutamine Metabolism and Induce Senescence of Hepatic Stellate Cells to Exert Anti-Hepatic Fibrosis Mechanism. Immunol Disord Immunother. 8:135
Received: 25-Dec-2022, Manuscript No. IDIT-23-21548; Editor assigned: 27-Dec-2022, Pre QC No. IDIT-23-21548 (PQ); Reviewed: 12-Jan-2023, QC No. IDIT-23-21548; Revised: 18-Jan-2023, Manuscript No. IDIT-23-21548 (R); Accepted: 01-Jan-0001 Published: 25-Jan-2023 , DOI: 10.35248/2593-8509.23.8.135
Copyright: © 2023 Duan X, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited