Journal of Antivirals & Antiretrovirals
Open Access
ISSN: 1948-5964
ISSN: 1948-5964
Research Article - (2021)Volume 13, Issue 2
Background: Coronavirus Disease 2019 (COVID-19) is a health crisis throughout the world. The widely used Real-time Reverse Transcriptase Polymerase Chain Reaction (rRT-PCR) method is most capable of describing the patient’s condition. Comorbidities can make patients more critical.
Methods: In this study, we shed light on the low Cycle Threshold (Ct) value of the N gene in the rRT-PCR test of the COVID-19 patients who had comorbidities, cure rate, and the needfulness of ICU (Intensive Care Unit) management. We had conducted the research in the Molecular Biology Laboratory of Chittagong Medical College between May and August 2020, then took the telephone interview with 300 positive patients who fulfilled the study criteria. We applied cluster-based logistic regression to analyze the data.
Results: Low Ct value of the N gene found 1.324 times more in Type 2 DM patients and 1.871 times higher in hypertensive patients, and hospitalized patients are 2.480 times more vulnerable to shift in ICU.
Conclusion: While infection with Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) frequently causes severe diseases, suspected cases with comorbid conditions should go through the rRT-PCR as early as possible.
Comorbidity; COVID-19; rRT-PCR; Ct value; N gene; Cluster-based logistic regression
SARS-CoV-2: Severe Acute Respiratory Syndrome Coronavirus 2; COVID-19: Coronavirus Disease 2019; Ct: Cycle Threshold; N: Nucleocapsid; rRT-PCR: Real-time Reverse Transcriptase Polymerase Chain Reaction; ICU: Intensive Care Unit; ICTV: International Committee on Taxonomy of Viruses; CoVs: Coronaviruses; HCoV: Human Coronaviruses; SARS-CoV: Severe Acute Respiratory Syndrome Coronavirus; MERS-CoV: Middle East Respiratory Syndrome Coronavirus; CDNA: Complementary DNA; RdRP: RNA-Dependent RNA Polymerase Gene; ORF1ab: Open Reading Frame 1ab; DM: Diabetes Mellitus; COPD: Chronic Obstructive Pulmonary Disease; CKD: Chronic Kidney Disease; IHD: Ischemic Heart Disease; CVD: Cardio-Vascular Disease; Exp(B): Odds Ratio; CI: Confidence Interval.
In December 2019, SARS-CoV-2 presented for the reason of Coronavirus Disease 19 (COVID-19) pandemic [1,2]. The International Committee on Taxonomy of Viruses (ICTV) declared this virus as the novel coronavirus in January 2020. ICTV named this virus because of their 9-12 nm long surface spikes that resemble a corona (equal to the crown in Latin). Coronavirus is an enveloped virus. They are single-stranded and belong to a positive- sense RNA genome. It is the 7th coronavirus and 3rd zoonotic Human Coronavirus (HCoV) of the century. It is posing threats to international health too.
It has now affected almost all countries and has led to over 25 million confirmed cases. Globally 870497 deaths have occurred till August 2020. There are also asymptomatic or mildly symptomatic cases, but enormous patients progress to severe COVID-19 with high mortality rates [1-4]. By July 2020, over 600,000 deaths recorded worldwide. Presently seven CoVs (including SARS- CoV-2) can cause human respiratory diseases to date, only Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV), Middle East Respiratory Syndrome Coronavirus (MERS-CoV), and SARS-CoV-2 have caused a large outbreak with high mortality. They can harm almost all systems: respiratory, gastrointestinal, cardiovascular, and nervous systems with variable severity of our body [5].
High viral loads in severe COVID-19 show higher viral replication and pathology, which results in multi-organ failure [6,7]. But it is not always true, as some studies found asymptomatic individuals can have high viral loads [8]. In this research, we explored the association of low Ct value of viral N gene with the comorbidities in both home and hospital cluster. We also tried to find the clinical outcomes of the corresponding patient in this research.
The molecular biology laboratory of Chittagong Medical College conducted this cross-sectional study from May 2020 to August 2020. Reverse Transcriptase-Polymerase Chain Reaction was done in the QuantStudio 5 Real-Time PCR instrument (96-Well 0.2 ml Block) by Sansure Biotech Novel Coronavirus Nucleic Acid Diagnostic Kit.
We included rRT-PCR confirmed COVID-19 patients who had three days of symptoms. Interviewers collected the following parameters of the corresponding patients over the phone: presence of symptoms, days of illness, contact with the symptomatic patient, types of personal protective equipment used, traveling history, oxygen requirement, hospital admission history, ICU support, co-morbidities, pH, HCO3, Oxygen saturation, Arterial CO2, Arterial O2, other baseline investigations, and treatment sheet.
The WHO website has provided several Real-time RT-PCR protocols for detecting SARS-CoV-2 in different countries [9]. Three fluorescent signals from FAM, ROX, and CY5 channels detected the viral copy DNA (cDNA). We selected the nucleocapsid protein gene N and the RNA-dependent RNA polymerase gene (RdRP) [also reported as Open Reading Frame 1ab (ORF1ab)] [10]. Internal control helped to check for biases of the total procedure. We used a positive control such as plasmids containing the complete SARS-CoV- 2 N gene in our study [11].
Here, the Ct values of the targeted genes express the RNA virus. We listed 300 patients from 436 COVID-19 patients in our study. We got relevant history properly from those 300 patients. We filled up the questionnaires by clinical characteristics and treatment of the patients. We took written informed consent from the patients or by next-of-kin. This study got approval from local regulatory authorities.
We used descriptive statistics, logistics regression, and cluster-based logistics regression to explain the N gene-related factors.
We classified patients' conditions into three categories: ICU support with non-invasive procedures, ICU support with mechanical ventilation, and those who need none of these. The patients who had a low Ct value (15.00-30.00) of the nucleocapsid gene needed more ICU support (13%) than the patients who had a high Ct value (30.01-40.00). With the ORF1ab gene, it was different. Here, patients with a high Ct value used 16.3% ICU (both Non-invasive and Mechanical ventilation) facilities than the patients with low Ct value. In our study, elderly patients (≥ 60 years age) belonging low (15.00-30.00) Ct value of N gene needed the highest (34.6%) ICU support. Children did not require any intensive care support. Only 40-59 years of age group patients needed mechanical ventilation (8.3%).
From Figure 1, we found the highest number (79) of cases who were suffering from hypertension (63.7%) had low Ct value (15.00-30.00) of N gene, and those patients needed more intensive care support for deteriorating their health conditions than the patients of high Ct value (30.01-40.00). Diabetic patients were in the second (48.4%) position. The patients who had low risk were suffering from CVD (2.4%) with a low Ct value of the N gene. From those comorbid conditions, hypertension and type 2 DM are significant with N gene values by chi-square test.
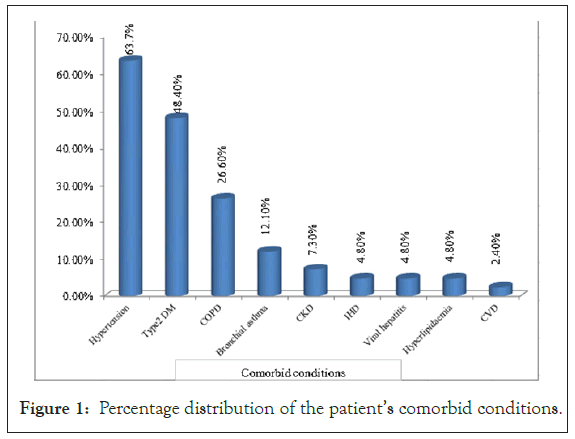
Figure 1: Percentage distribution of the patient’s comorbid conditions.
Table 1 represented that home treatment cured the patients who were not suffering from type 2 DM (29%) and hypertension (25%), though they had a low Ct value of the N gene. But diabetic and hypertensive patients needed more (11.00% and 12.00%, respectively) hospital treatment for those who had a low Ct value of the N gene. Having a high Ct value of the N gene, non-diabetic and normotensive patients cured more in-home treatment (25.00% and 25.70%, respectively).
| Comorbidity | Categories | Low Ct (15.00-30.00) of N gene | High Ct (30.01-40.00) of N gene | ||
|---|---|---|---|---|---|
| Home | Hospital | Home | Hospital | ||
| Type 2 DM | No | 29.00% | 17.00% | 25.00% | 9.00% |
| Yes | 3.00% | 11.00% | 2.00% | 4.00% | |
| Hypertention | No | 25.00% | 16.00% | 25.70% | 7.00% |
| Yes | 7.00% | 12.00% | 1.30% | 6.00% | |
Table 1: N gene range with cluster-based comorbid conditions.
Building logistics regression and cluster-based logistic regression, we defined the N gene range (low and high) as a dependent variable and significant comorbidities as independent variables. In the logistics regression model for general data, type 2 diabetic patients had a 1.324 times more low Ct value of the N gene. Similarly, in the home cluster, it was 1.192 times higher with CI (0.365, 3.888). But in the hospital cluster, the scenario was reversed (0.605), which was not significant. Hypertensive patients possessed 1.871 times significantly (p=0.042) more low Ct value of the N gene in the full dataset. Similarly, it was 1.375 times more in the hospital cluster. But, the home cluster represented that hypertensive patients had significantly (0.179 times with p-value 0.003) low Ct value of N gene (Table 2).
| Logistics regression model for general data | |||||||
|---|---|---|---|---|---|---|---|
| Comorbidities | Categories | B | S.E. | Sig. | Exp(B) | 95% C.I | |
| Lower | Upper | ||||||
| Type 2 DM | Yes | 0.28 | 0.337 | 0.046 | 1.324 | 0.683 | 2.565 |
| No | - | - | - | 1 | - | - | |
| Hypertention | Yes | 0.626 | 0.308 | 0.042 | 1.871 | 1.024 | 3.419 |
| No | - | - | - | 1 | - | - | |
| Constant | -1.105 | 0.315 | 0 | 0.331 | |||
| Cluster-based logistics regression (home cluster) | |||||||
| Variable | Categories | B | S.E. | Sig. | Exp(B) | 95% C.I | |
| Lower | Upper | ||||||
| Type 2 DM | Yes | 0.175 | 0.603 | 0.071 | 1.192 | 0.365 | 3.888 |
| No | - | - | - | 1 | - | - | |
| Hypertention | Yes | -1.718 | 0.582 | 0.003 | 0.179 | 0.057 | 0.562 |
| No | - | - | - | 1 | - | - | |
| Constant | 0.016 | 0.166 | 0.923 | 1.016 | |||
| Cluster-based logistics regression (hospital cluster) | |||||||
| Variable | Categories | B | S.E. | Sig. | Exp(B) | 95% C.I | |
| Lower | Upper | ||||||
| Type 2 DM | Yes | -0.503 | 0.448 | 0.262 | 0.605 | 0.251 | 1.456 |
| No | - | - | - | 1 | - | - | |
| Hypertention | Yes | 0.319 | 0.425 | 0.045 | 1.375 | 0.598 | 3.162 |
| No | - | - | - | 1 | - | - | |
| Constant | -0.733 | 0.273 | 0.007 | 0.48 | |||
Table 2: Cluster-based logistic model: Effect of comorbidities on low Ct value.
From Table 3, 23.00% of patients needed ICU, where 20.00% used non-invasive methods, and 3.00% went for mechanical ventilation. Besides, 11.00% of hospitalized patients needed ICU, who had a low Ct value of the N gene. On the alternative, 8.00% of hospitalized patients needed ICU, who had a high Ct value of the N gene.
| ICU | Low Ct (15.00-30.00) of N gene | High Ct (30.01-40.00) of N gene | ICU support type | |||
|---|---|---|---|---|---|---|
| Home | Hospital | Home | Hospital | Non-invasive | Mechanical ventilation | |
| No | 30.00% | 17.00% | 25.00% | 5.00% | 20.00% | 3.00% |
| Yes | 2.00% | 11.00% | 2.00% | 8.00% | ||
Table 3: ICU support according to Ct value range of N gene.
Building logistics regression and cluster-based logistic regression, we defined the patients' condition into two categories: Who needed ICU support and those who did not need any ICU support rolled as a dependent variable. Table 4 represented that ICU patients were 0.17 times lower than those who had a low Ct value (15.00-30.00) of viral N gene in the rRT-PCR method than the high Ct value (30.01-40.00). But, hospital clusters in the cluster-based logistic regression method, ICU admissions were significantly 2.480 times (CI 0.882, 5.026) higher than patients having a low Ct value of the N gene.
| Logistics regression model for general data | |||||||
|---|---|---|---|---|---|---|---|
| Target | Categories | B | S.E. | Sig. | Exp(B) | 95% C.I | |
| Lower | Upper | ||||||
| N Gene | Low | -0.187 | 0.278 | 0.052 | 0.83 | 0.481 | 1.43 |
| High | - | - | - | 1 | - | - | |
| Constant | -1.099 | 0.211 | 0 | 0.333 | - | - | |
| Cluster-based logistics regression (home cluster) | |||||||
| Categories | B | S.E. | Sig. | Exp(B) | 95% C.I | ||
| Lower | Upper | ||||||
| N Gene | Low | -0.182 | 0.598 | 0.076 | 0.833 | 0.258 | 2.691 |
| High | - | - | - | 1 | - | - | |
| Constant | -2.526 | 0.424 | 0 | 0.08 | - | - | |
| Cluster-based logistics regression (hospital cluster) | |||||||
| Target | Categories | B | S.E. | Sig. | Exp(B) | 95% C.I | |
| Lower | Upper | ||||||
| N Gene | Low | -0.905 | 0.398 | 0.023 | 2.48 | 0.882 | 5.026 |
| High | - | - | - | 1 | - | - | |
| Constant | 0.47 | 0.329 | 0.153 | 1.6 | - | - | |
Table 4: Cluster-based logistic model: Effect of N gene on ICU support.
We found that patients with low Ct values were more likely to have Hypertension (n=79), Type 2 DM (n=60), COPD (n=33), Bronchial Asthma (n=15), CKD (n=9). Decreased oxygenation in the heart and kidney or increased infection may be responsible for this. Another study revealed that SARS-CoV-2 directly infects both the heart and kidney [12]. Renal and cardiovascular complications are common in severe COVID-19 [13,14]. Further studies can evaluate the relationships between the lower level of Ct value of the N gene and disease severity.
Maximum patients of COVID-19 had mild symptoms, but 18% to 33% of patient’s required invasive procedures in ICU, and 20% of patients died in the hospital in different studies [15-20]. Previous studies reflect that the low Ct value corresponds with the high viral loads. Our study highlights the low level of the Ct value of the N gene detecting SARS-CoV-2 can deteriorate patients' condition.
Viral loads express COVID-19 severity as a prognostic biomarker, but there is no harmony across the studies [21-24]. In our analysis, more patients in ICU had low Ct of N gene. Magleby et al. also found a similar result [21]. Ct values help to identify patients who are in a risky zone and who need more intensive care. It may also help to select antiviral agents [25].
There were some limitations in our study. First, we analyzed only the Ct value of a single time sample, so we could not assess viral load dynamics overtime. Second, our study population size was small, and the study period was short.
We pointed the hazardous conditions of the patients depend on the Ct value range of the N gene. Even it is worst when there are comorbidities (Type 2 DM and Hypertension). This information is useful for selecting patients for COVID-19 who require precise management and follow-up at home or hospital. These findings also highlight the critical role of viral load. It also suggests that clinicians should follow the Ct values in identifying patients at high-risk conditions.
Ethics approval and consent to participate
The study protocol was assessed and approved by the medical ethics committee of the Chittagong Medical College and Hospital, Chattogram.
Consent for publication: Not applicable.
Availability of data and materials: Upon reasonable request.
Competing interests: The authors declare no conflict of interest.
Funding: Not applicable.
Author contributions
Sujan Rudra: Data curation; Statistical analysis.
Shuva Das: Conceptualization, roles/writing-original draft.
Md. Ehsanul Hoque: Supervision.
Abul Kalam: Writing, review and editing.
Mohammad Arifur Rahman: Validation, visualization.
Swagata Nandy Shizuka: Investigation.
Tazrina Rahman: Methodology.
All the authors have accepted equal responsibility for the entire content of this submitted manuscript and approved submission.
We would like to thanks the editors and referees whose constructive criticism led us to develop the presentation and maintain the quality of the paper. We would also like to thanks the Molecular Biology Laboratory of Chittagong Medical College, Chattogram, to give their almost all opportunity for conducting the research.
Citation: Rudra S, Das S, Hoque ME, Kalam A, Rahman MA, Shizuka SN, et al. (2021) Comorbidities of COVID-19 Patients with Low Cycle Threshold (Ct) Value of Nucleocapsid (N) Gene: An Application to Cluster-Based Logistic Model. J Antivir Antiretrovir. 13:214.
Received: 29-Jan-2021 Accepted: 12-Feb-2021 Published: 19-Feb-2021 , DOI: 10.35248/1948-5964.21.13.214
Copyright: © 2021 Rudra S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.