Clinical Pediatrics: Open Access
Open Access
ISSN: 2572-0775
ISSN: 2572-0775
Research Article - (2023)Volume 8, Issue 3
Background: Y-RNAs are small noncoding RNAs firstly identified in patients suffering the Sjogren’s syndrome and lupus erythematosus, having different predicted cellular functions. Y-RNAs are very abundant ncRNAs present in serum and only a few is known respect their molecular roles.
Objective: The aim of the study was to determine the circulating Y-RNAs expression from blood serum of pediatric patients with pilocytic or diffuse astrocytoma.
Materials and methods: Y-RNAs expression was determined by means of the Human Transcriptome Array (HTA) 2.0 arrays. In addition, a bioinformatic approximation of the possible biological functions of Y-RNAs was determined with the Random Forest (RF) and Vector Support Machine (VSM) algorithms.
Results: Data showed a differential expression of RNY-3, RNY-4, and RNY-5 in pediatric patients with astrocytoma, relative to the control. Meanwhile, RNY-1 was shown to be upregulated in the diffuse condition versus the pilocytic. The bioinformatic analysis showed that RNY-4 and RNY-5 had the highest scores of interaction with Claudin, Toll-like receptors, and Hyaluronidase-1. In addition, the Y-RNAs loop domain by itself had the highest score of interaction with B cell receptor CD22 (RNY-3) and Toll-like receptor 7 and 3 (RNY-4).
Conclusion: Our results showed, for the first time a differential expression of circulating Y-RNAs in pediatric astrocytoma, allowing to distinguish pediatric subjects without cancer from patients with pediatric diffuse or pilocytic astrocytoma and their potential involvement in regulating diverse biological processes, such as immune activation-suppression, cell signaling, selective transcription, and cell proliferation through activation of DNA replication.
Toll-like receptors; Exosome; Regulation; Pediatric
The RNA-Protein interactions have a critical participation in the cell function regulation [1,2]. It is estimated nearly 6% from the human proteome could bind to RNA. Evidence is available of RNA-proteins network interactions regulating cell signaling and genomic expression [2,3]. Around 98% of DNA does not code for proteins [4]. Approximately 99% from the total RNA, in a mammal cell, belongs to non-coding RNA (ncRNA) [5]. The ncRNAs are classified according to their length as small non-coding RNA (sncRNAs) with less than 200 nucleotides (nt) and long non-coding RNA (lncRNAs) with 200 or more nucleotides on length. A type of sncRNA are Y-RNAs, which were discovered by forming ribonucleoprotein (RNPs) complexes with the Ro60 (Ro60, Y-RNA Binding Protein) and La (Sjogren syndrome type B antigen (SS-B)) proteins, and autoantigens in patients suffering the Sjogren’s syndrome and lupus erythematosus [6]. Y-RNAs, so called due to their identification in the cell cytoplasm (Y for cytoplasmic), are a family of RNAs transcribed by the RNA polymerase III, defined by their structure and affinity by the Ro60 protein [7]. The four genes are located as a tandem set at the chromosomal locus 7q36 [7]. But each Y-RNA has its own promoter and is independently transcribed [8].
Structurally, according to Kowalski and Krude [9], human Y-RNAs have four domains the 5' and 3' ends of the Y-RNAs hybridize forming a lower and upper double chain, an intermediate loop, and a polyuracil tail. The conserved binding sites of the Ro60 and La proteins are located in the lower double chain and in the polyuracil tail, respectively. The middle loop is the most variable region and there is evidence that it binds to various proteins, including nucleolin [10].
Y-RNAs are highly conserved molecules and their translocation from the nucleus to the cytoplasm requires their binding to the Ro60 protein and to other snRNAs [11-13]. Y-RNAs can be secreted inside Extracellular Vesicles (EVs) derived from blood or stem cells, immune system cells, or tumor cells [14-17].
The first analyses concerning Y-RNA quantification in human cancers were performed with solid tumors from carcinomas and adenocarcinomas of the lung, kidney, bladder, prostate, colon, and cervix. As a general rule, all four Y-RNAs were shown to be overexpressed in this type of cancers [18]. However, the RNY-5 expression does not generally correlate with that of the other, which could be explained by the fact that its location (cell nucleus) is different from that of the others (cell cytoplasm) [19]. According to this, a recent research has reported the downregulation of all human Y-RNAs in breast cancer, but RNY-1, RNY-3, and RNY-4 showed a high expression correlation, whereas that of RNY-5 was less correlated [20]. Cancer-derived Extracellular Vesicles (Evs) in myelogenous leukemia have a relatively high amount of RNY-5, which is the most abundant RNA after rRNA (ribosomal RNAs) and tRNA (transfer RNAs) [21]. In addition, evidence suggests a role for Extracellular Vesicles (EVs) as regulators for brain tumor progression [22].
Astrocytomas (Ast) are a heterogeneous group of the central nervous system tumors that differ in their morphologic characteristics and localization. Until 2016, pediatric and adult astrocytomas were classified into four main groups based on specific criteria established by the World Health Organization (WHO). These groups included, pilocytic Astrocytomas (PAst; grade I), diffuse Astrocytomas (DAst; grade II), and anaplastic Astrocytomas (AAst; grade III), as well as Glio-blastoma Multiforme (GBM; grade IV) [23]. pAst and dAst are the most frequent tumors in children and aAst and GBM are relatively rare tumors [24]. Pediatric Ast (p-Ast) are the leading cause of childhood solid tumor deaths and although p-Ast and adult Ast (a-Ast) share signaling pathways that are altered, the molecular components are distinct [25-27]. Based on this, P-Ast and a-Ast are different clinical entities with specific genetic alterations. In 2016, the WHO proposed a new Central Nervous System (CNS) tumor classification based on molecular and epigenetic markers, as well as in the clinical behavior of adult patients; however, more studies are needed to provide more molecular tools to this new classification with the aim of having a better Ast classification, impacting the diagnosis, prognosis, patient’s survival, and drug therapy. In the present work, the circulating Y-RNAs expression was determined by means of the Human Transcriptome Array (HTA) 2.0 Arrays from blood serum samples from pediatric patients with Ast. Bioinformatical analysis showed their interaction with different proteins, suggesting their involvement mainly in the control of immune system, cell signaling, and DNA replication.
Samples
Blood serum samples from pediatric patients with Ast (PAst, n=10, four boys and six girls; DAst, n=13, five boys and eight girls) were collected from 2017 to 2020. All children included in this study were patients from the Children's Hospital Dr. Silvestre Frenk Freund, National Medical Center XXI Century, Mexican Institute of Social Security (IMSS). The patient’s age range was 0-15 years with no history of relatives with cancer. All children’s relatives or tutors signed an informed consent.
Ethics
The project had the approval of the Children's Hospital Dr. Silvestre Frenk Freund, National Medical Center XXI Century, Mexican Institute of Social Security, Ethics in Security Research (R-2014-3603-27), which is in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Exosome and RNA isolation
Exosomes were isolated with the total isolation exosome kit (from serum) (ThermoScientific) and the presence and integrity of exosomes was visualized by electron microscopy. For RNA isolation, exosomes were incubated for 15 h (4°C) with TRIZOL® (this was to optimally resuspend exosomes) and later with 40 µL chloroform for 15 minutes at 4°C. The aqueous phase (RNA) was transferred to a new tube and 500 µL isopropanol were added for RNA precipitation. Finally, RNA was washed three times with 75% ethanol and resuspended in 5 µL RNAse-free water. RNA quantification was performed by spectrophotometry (Nanodrop 1000, Thermo Fisher Scientific).
GeneChip® human transcriptome array 2.0
Three conditions (Control (n=50), pilocytic (n=10), and diffuse (n=15)) were used for the analysis of differential expression of the exosome-derived transcriptomes. The Human Transcriptome Array (HTA) 2.0 (ThermoScientific) determined the differential expression among arrays of distinct experimental conditions. The expression analysis was done by groups (Pulls) and not individually, therefore, differences in the transcript expression for each sample could not be detected. With this array, it was possible to evaluate >285,000 full-length covered transcripts, >245,000 coding transcripts, >40,000 non-coding transcripts, and >339,000 probe sets covering exon-exon junctions. Median expression levels (p<0.001) were significant. Data were uploaded to GEO (Gene Expression Omnibus; number: GSE216815).
Bioinformatic analyses
To perform bioinformatic analyses, the web server interface (free access), belonging to the Iowa State University, was used (RNA-Protein Interaction Prediction Server Protein-RNA Interaction Database (PRIDB) RPI-Seq Version Dobbs Lab) [28,29].
Sequences of the four human Y-RNAs were downloaded from the National Center for Biotechnology Information (NCBI) nucleotide databases.
Protein sequences of cell receptors from diverse protein families were obtained by using key words and Homo sapiens (txid9606), being in total 1319 sequences. Filtered applied: from zero to twelve hundred amino acids in length. Protein sequences of toll-like cell receptors were downloaded from the NCBI protein database by using key words: (Toll, Cell, Receptors, and Human) and Homo sapiens (txid9606), being in total one hundred and nineteen not filtered sequence. The removal of the terminal segments in the 5´and 3´terminals, to generate transcripts only with the loop domain from the four human Y-RNAs, was performed.
Protein sequences of the Wnt cell receptors were downloaded by using key words and Homo sapiens (txid9606), being in total thirty-seven sequences. In the same way, protein sequences related to the Origin of Replication Complex (ORC) were retrieved using key words and Homo sapiens (txid9606), being in total one thousand, two hundred eighty-six. Filtered applied from zero to one thousand two hundred amino acids in length. All the sequences were checked manually to avoid redundant annotations or duplicated sequences and formatted and ready to be entered into the web server: http://pridb.gdcb.iastate.edu/RPISeq.
In this work, data obtained with the Random Forest (RF) algorithm were mainly discussed, since its predictions are the most validated at the experimental level. Only in the interactions of Y-RNAs with the proteins related to the Origin of Replication Complex (ORC), the scores obtained with Random Forest (RF) and SVM were shown. This was with the purpose of comparing the results obtained with both classifiers. Although values greater than 0.5 were taken as significant, those closest to 1 have the highest probability of occurring in vivo. Essentially, RPIseq exploits the aminoacidic composition of proteins sequences and the ribonucleotides composition from the RNA sequences to predict the probability of in vivo interactions of a pair (RNA-Protein). This web server RPIseq implements the RPIseq method developed by Muppirala [28,29].This algorithm takes a data pair of sequence belonging to RNA and protein as an input, and computes probabilities of interaction through the Random Forest (RF) and SVM trained classifiers using the datasets from the RPI2241. This interface could accept many proteins against a specific RNA molecule or vice versa users could introduce a maximum of 100 sequences.
String protein network
Cytoscape is an open source software platform for visualizing molecular interaction networks and biological pathways and integrate these networks with annotations, gene expression profiles, and other state data. Although Cytoscape was originally designed for biological research, now it is a general platform for complex network analysis and visualization [30]. The String Networks were constructed with Cytoscape 3.8.2 for windows, using String Protein Query database.
Enrichment analyses
We used PANTHER GO-slim", which uses a selected set of terms from the Gene OntologyTM for classifications by molecular function, biological process, and cellular component. The PANTHER Protein Class ontology was adapted from the PANTHER/X molecular function ontology and includes commonly used classes of protein functions, many of which are not covered by Gene Ontology (GO) molecular function. Download the classes and relationship information [31].
GeneChip HTA 2.0 array
The comparison between the Pilocytic and Control conditions showed subtle changes in the expression of RNY-3 (upregulated in PAst) and RNY-4 and -5 (both downregulated) as shown in Figure 1A and Table 1. By contrast, RNY-1, -3, and -5 were highly upregulated in the diffuse condition compared to the Pilocytic and Control conditions as shown in Figures 1B and 1C and Table 1.
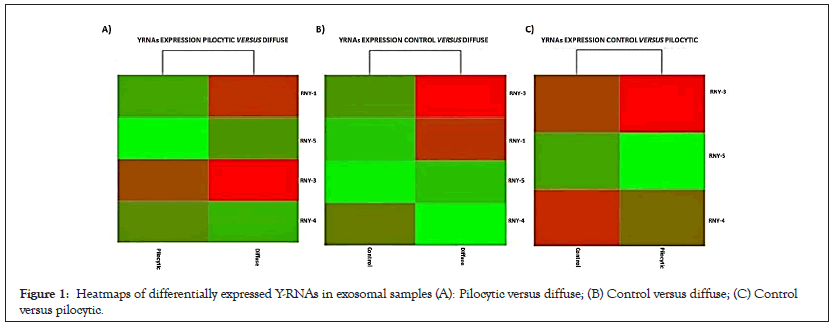
Figure 1: Heatmaps of differentially expressed Y-RNAs in exosomal samples (A): Pilocytic versus diffuse; (B) Control versus diffuse; (C) Control versus pilocytic.
| Y-RNA | Diffuse | Pilocytic | Fold change |
|---|---|---|---|
| RNY-1 | 15.95 | 9.37 | 95.69 |
| RNY-5 | 10.18 | 5.5 | 25.63 |
| RNY-3 | 18.7 | 14.19 | 22.68 |
| RNY-4 | 8.76 | 10.68 | -3.77 |
| Y-RNA | Diffuse | Control | Fold change |
| RNY-3 | 18.74 | 11.58 | 143.58 |
| RNY-1 | 16.15 | 9.53 | 98.05 |
| RNY-5 | 9.81 | 8.09 | 3.29 |
| RNY-4 | 7.67 | 12.7 | -32.63 |
| Y-RNA | Pilocytic | Control | Fold change |
| RNY-3 | 14.43 | 11.58 | 7.21 |
| RNY-5 | 5.22 | 8.09 | -7.3 |
| RNY-4 | 10.19 | 12.7 | -5.68 |
Table 1: Y-RNA relative levels detected in exosomal samples of pediatric astrocytoma and controls.
Predicted human Y-RNAs interactions with proteins
To gain insight in Y-RNAs knowledge, the interaction of Y-RNAs with cell receptors was bioinformatically studied in most cases, the Random Forest (RF) algorithm showed the highest scores of interactions for RNY-4 and RNY-5, followed by RNY-1 and RNY-3 (Table 2).
| Protein | Y-RNAs Subtypes | |||||
|---|---|---|---|---|---|---|
| NP_ or XP_ | 1 | 3 | 4 | 5 | A | |
| Higher average score for each human Y-RNAs with different families of receptor proteins | ||||||
| Transient receptor potential cation channel subfamily M | 1353082 | 0.7 | 0.75 | 0.85 | 0.85 | 0.7875 |
| G-protein coupled receptor 37-like 1 | 4758.3 | 0.75 | 0.65 | 0.85 | 0.9 | 0.7875 |
| Claudin domain-containing protein 1 isoform a | 1035271 | 0.75 | 0.7 | 0.95 | 0.7 | 0.775 |
| Lysosome membrane protein 2 isoform 1 | 5497.1 | 0.7 | 0.85 | 0.85 | 0.7 | 0.775 |
| Hyaluronidase-1 isoform 3 | 695015.1 | 0.6 | 0.7 | 0.85 | 0.95 | 0.775 |
| Sorting nexin-4 | 3785.1 | 0.75 | 0.6 | 0.85 | 0.85 | 0.7625 |
| Linker for activation of T-cells family member 1 isoform a | 55202.1 | 0.75 | 0.6 | 0.85 | 0.85 | 0.7625 |
| Fibroblast growth factor 23 | 65689.1 | 0.65 | 0.8 | 0.85 | 0.75 | 0.7625 |
| Claudin domain-containing protein 1 isoform d | 1035290 | 0.65 | 0.7 | 0.9 | 0.8 | 0.7625 |
| Tumor necrosis factor receptor superfamily member 19 | 61117.2 | 0.75 | 0.7 | 0.85 | 0.75 | 0.7625 |
| Higher average score for each of the human Y-RNAs with Toll-like proteins | ||||||
| B-cell receptor CD22 isoform 1 | 1762.2 | 0.7 | 0.65 | 0.9 | 0.75 | 0.75 |
| Protein unc-93 homolog B1 | 112192.2 | 0.65 | 0.7 | 0.75 | 0.85 | 0.7375 |
| Toll-like receptor 8 isoform 2 | 619542.1 | 0.6 | 0.8 | 0.9 | 0.65 | 0.7375 |
| Toll-like receptor 7 | 57646.1 | 0.55 | 0.65 | 0.9 | 0.8 | 0.725 |
| Toll-like receptor 10 isoform a | 1017388 | 0.55 | 0.65 | 0.8 | 0.9 | 0.725 |
| Toll-like receptor 8 isoform 1 | 57694.2 | 0.65 | 0.7 | 0.9 | 0.65 | 0.725 |
| Toll-like receptor 10 isoform a | 112218.2 | 0.55 | 0.65 | 0.8 | 0.9 | 0.725 |
| Toll-like receptor 5 | 3259.2 | 0.55 | 0.6 | 0.95 | 0.8 | 0.725 |
| MHC class I-related gene protein isoform 1 | 1522.1 | 0.65 | 0.7 | 0.85 | 0.65 | 0.7125 |
| Interleukin-1 receptor type 1 isoform 4 | 1307913 | 0.75 | 0.55 | 0.85 | 0.7 | 0.7125 |
| Higher average score for each of the human Y-RNAs Loop domains with Toll-like proteins | ||||||
| B-cell receptor CD22 isoform 1 | 1762.2 | 0.7 | 0.9 | 0.8 | 0.75 | 0.7875 |
| B-cell receptor CD22 isoform 3 | 1172029 | 0.7 | 0.85 | 0.8 | 0.7 | 0.7625 |
| Toll-like receptor 7 | 57646.1 | 0.6 | 0.75 | 0.85 | 0.8 | 0.75 |
| Toll-like receptor 3 | 3256.1 | 0.65 | 0.7 | 0.85 | 0.7 | 0.725 |
| Interleukin-1 receptor type 1 isoform 1 | 868.1 | 0.6 | 0.8 | 0.8 | 0.65 | 0.7125 |
| B-cell receptor CD22 isoform 4 | 1172030 | 0.6 | 0.8 | 0.75 | 0.7 | 0.7125 |
| Interleukin-1 receptor type 1 isoform 4 | 1307913 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 |
| Protein unc-93 homolog B1 | 112192.2 | 0.65 | 0.7 | 0.7 | 0.75 | 0.7 |
| Toll-like receptor 10 isoform a | 1017388 | 0.6 | 0.75 | 0.7 | 0.7 | 0.6875 |
| Single Ig IL-1-related receptor | - | 0.55 | 0.75 | 0.7 | 0.75 | 0.6875 |
| Higher average score for each of the human Y-RNAs against wnt cell rceptors | ||||||
| Alpha-tubulin N-acetyltransferase 1 isoform 1 | 1026892 | 0.6 | 0.5 | 0.65 | 0.6 | 0.5875 |
| Glycogen synthase kinase-3 beta isoform 2 | 1139628 | 0.55 | 0.55 | 0.6 | 0.4 | 0.525 |
| Alpha-tubulin N-acetyltransferase 1 isoform 3 | 1177653 | 0.55 | 0.55 | 0.7 | 0.5 | 0.575 |
| Metastasis-associated protein MTA1 isoform MTA1s | 1190187 | 0.6 | 0.6 | 0.75 | 0.55 | 0.625 |
| CCN family member 4 isoform 3 | 1191798 | 0.4 | 0.5 | 0.65 | 0.6 | 0.5375 |
| CCN family member 4 isoform 4 | 1191799 | 0.4 | 0.45 | 0.6 | 0.55 | 0.5 |
| Alpha-tubulin N-acetyltransferase 1 isoform 4 | 1241881 | 0.6 | 0.55 | 0.65 | 0.55 | 0.5875 |
| Protein Wnt-8a isoform 1 precursor | 1287867 | 0.5 | 0.55 | 0.8 | 0.75 | 0.65 |
| Protein Wnt-8a isoform 2 precursor | 1287868 | 0.45 | 0.55 | 0.7 | 0.7 | 0.6 |
| Alpha-tubulin N-acetyltransferase 1 isoform 5 | 1305691 | 0.6 | 0.55 | 0.65 | 0.55 | 0.5875 |
Table 2: Top 10 interactions of human Y-RNAs.
RNY-4 and RNY-5 showed most interaction with Claudin domain-containing protein, G-protein coupled receptor 37, cation channel subfamily M, Hyaluronidase-1, and Sorting nexin-4, among others. Meanwhile RNY-1 with G-protein coupled receptor 37 and Linker for activation of T-cells for example. RNY-3 has the potential to bind with Lysosome membrane protein 2 and Fibroblast growth factor 23.
Cell receptors
Results generated by the Random Forest (RF) algorithm showed high scores (7, 8.5, or 9) for the Y-RNAs and generalized cell receptors interactions. The most significant interactions corresponded to RNY-4 and RNY-5, followed by RNY-1 and RNY-3 as shown in Figures 2A-2C. RNY-4 downregulation in the pilocytic and diffuse conditions compared to the control might result in aberrant Toll-like Receptors (TLRs) activation or OCR control. This might be affecting cell division and response to stimulus, such as communication of immune related cells with the local environment as shown in Figures 2D-2F and Figures 3A-3C. Meanwhile, the overexpression of RNY-1 and RNY-3, in the diffuse and control conditions relative to the pilocytic one, might be affecting innate and acquired immune related process as well as cell signaling mechanisms (Figures 2A and 3A).
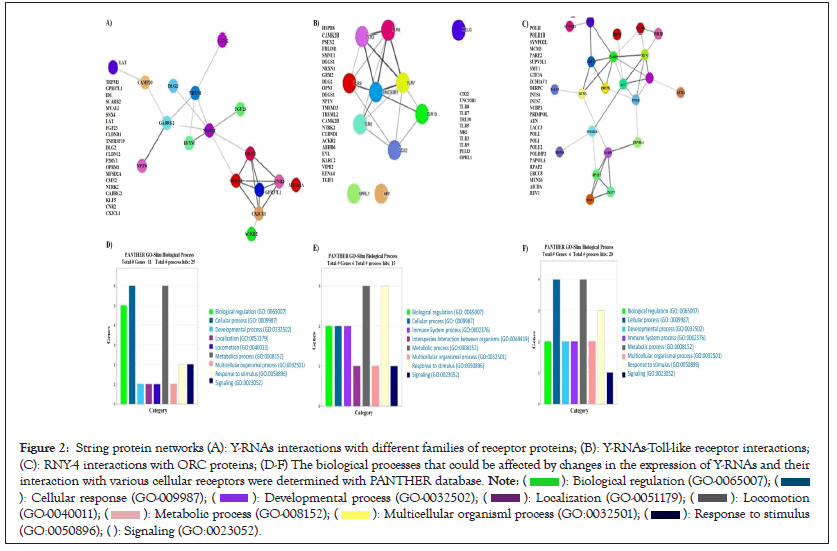
Figure 2: String protein networks (A): Y-RNAs interactions with different families of receptor proteins; (B): Y-RNAs-Toll-like receptor interactions;
(C): RNY-4 interactions with ORC proteins; (D-F) The biological processes that could be affected by changes in the expression of Y-RNAs and their
interaction with various cellular receptors were determined with PANTHER database. 
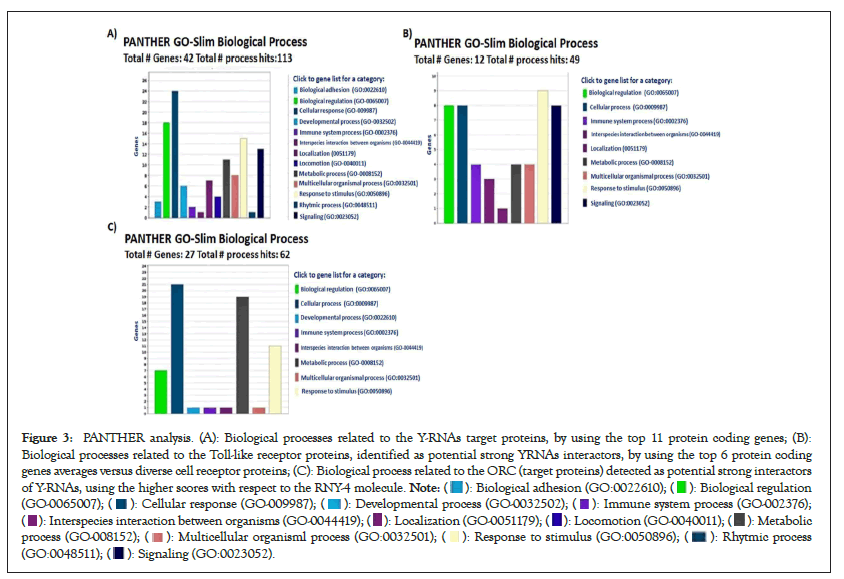
Figure 3: PANTHER analysis. (A): Biological processes related to the Y-RNAs target proteins, by using the top 11 protein coding genes; (B):
Biological processes related to the Toll-like receptor proteins, identified as potential strong YRNAs interactors, by using the top 6 protein coding
genes averages versus diverse cell receptor proteins; (C): Biological process related to the ORC (target proteins) detected as potential strong interactors
of Y-RNAs, using the higher scores with respect to the RNY-4 molecule. 
Toll-like receptors
Due to experimental evidence indicating the interaction of many Y-RNAs with the Toll-like Receptor 3 (TLR3) and Toll-like Receptor 7 (TLR7) receptors [28]. Y-RNAs-Toll-like receptors interactions were predicted. Like the above results, the RNY-4 and RNY-5 had the most significant score values of interaction (according to random forest), with a 0.9 score for the RNY-4 and RNY-5 interaction with CD22 (CD22 molecule) and TLR7 and TLR8. The RNY-4 interaction with TLR5 showed an interaction score of 0.95 and that of the RNY-5 with TLR10 and the unc-93 protein were 0.9 and 0.85, respectively. When we considered the average score for the four Y-RNAs, the highest score was with CD22, followed by unc-93, and TLRs 7 and 8. Interestingly, RNY-4 had the most significant score of interaction with TLRs. Y-RNAs interactions with TLRs might be related to innate immune system regulation and consequent feedback by cytokines and/or chemokines secretion by target cells or tissues.
Wnt receptors
To the best of our knowledge, Y-RNAs interactions with Wnt receptors have not been previously reported; therefore, these interactions were tested. Y-RNAs-Wnt receptors interactions showed very low score values, suggesting weak probabilities of interaction of these molecules in vivo.
Y-RNAs loop domain
Recent evidence shows the in vivo RNase 1 processing of Y-RNAs, producing different fragments of different size of these RNAs [29]. Based on this, the interaction of the loop domain of the Y-RNAs with TLRs was tested. Data showed high scores of interactions, which were very similar to that obtained when the whole Y-RNAs (with their four domains) were, analyzed (Table 2).
Y-RNAs interactions with ORC-associated proteins
Predictions showed some of the highest scores for Y-RNAs interactions with ORC-associated proteins, particularly for RNY-4 and RNY-5 as shown in Table 3. This reinforces previous evidence indicating the presence of Y-RNAs in Origin of Replication Complex (ORC) [32]. Interactions with the highest scores involved diverse proteins related to Deoxyribonucleic Acid (DNA) replication, i.e., DNA polymerases, transcription factors, helicases, and others. According to the Random Forest (RF) classifier, RNY-4 seems to be particularly important since it showed a lot of predicted interactions with this type of proteins. These interactions might have a biological impact on DNA replication and cell proliferation (Table 3).
| NP_ or XP_ | RF | SVM | |
|---|---|---|---|
| Top 10 interactions for RNY-1 with ORC related proteins | |||
| DNA polymerase eta isoform X2 | 24302234.1 | 0.8 | 0.887 |
| Domain-containing protein 1 | 60386.1 | 0.8 | 0.606 |
| General transcription factor 3C polypeptide 2 isoform b | 1030598.1 | 0.8 | 0.901 |
| Proline-rich nuclear receptor coactivator 1 | 6804.1 | 0.8 | 0.88 |
| DNA polymerase lambda isoform X6 | 11537958.1 | 0.75 | 0.689 |
| DNA polymerase lambda isoform X10 | 16871575.1 | 0.75 | 0.665 |
| TATA box-binding protein-associated factor RNA polymerase I subunit A | 647603.1 | 0.75 | 0.787 |
| General transcription and DNA repair factor IIH helicase subunit XPD | 391.1 | 0.75 | 0.787 |
| ATPase WRNIP1 isoform 2 | 569079.1 | 0.75 | 0.756 |
| General transcription factor 3C polypeptide 2 isoform a | 1305838.1 | 0.75 | 0.92 |
| Top 10 interactions for RNY-3 with ORC related proteins | |||
| DNA-directed RNA polymerase III subunit RPC5 isoform 5 | 1244965 | 0.8 | 0.968 |
| General transcription factor 3C polypeptide 2 isoform b | 1030598.1 | 0.8 | 0.984 |
| General transcription factor 3C polypeptide 2 isoform b | 1512.1 | 0.8 | 0.984 |
| F-box only protein 7 isoform 1 | 36311.3 | 0.8 | 0.961 |
| E3 ubiquitin-protein ligase Jade-2 isoform X3 | 24301775.1 | 0.8 | 0.96 |
| DNA polymerase eta isoform X4 | 24302235.1 | 0.75 | 0.965 |
| DNA polymerase lambda isoform X10 | 16871575.1 | 0.75 | 0.925 |
| DNA-directed RNA polymerase III subunit RPC5 isoform 1 | 60589.1 | 0.75 | 0.97 |
| DNA-directed RNA polymerase III subunit RPC5 isoform 3 | 1244964.1 | 0.75 | 0.98 |
| DNA-directed RNA polymerase III subunit RPC5 isoform 2 | 1244962.1 | 0.75 | 0.968 |
| Top 10 interactions for RNY-4 with ORC related proteins | |||
| DNA polymerase delta subunit 3 isoform 2 | 1350526.1 | 0.95 | 0.987 |
| DNA polymerase eta isoform X2 | 24302234.1 | 0.95 | 0.984 |
| DNA polymerase delta subunit 3 isoform X1 | 11543036.1 | 0.95 | 0.99 |
| DNA-directed RNA polymerase I subunit RPA2 isoform 5 | 1269705.1 | 0.95 | 0.989 |
| Synaptopodin 2-like protein isoform b | 79151.2 | 0.95 | 0.992 |
| DNA-directed RNA polymerase I subunit RPA2 isoform 6 | 1269708.1 | 0.9 | 0.99 |
| DNA replication licensing factor MCM3 isoform 6 | 1353301.1 | 0.9 | 0.98 |
| Poly [ADP-ribose] polymerase 2 isoform X1 | 5267304.1 | 0.9 | 0.974 |
| ATP-dependent RNA helicase SUPV3L1, Mitochondrial isoform 1 | 3162.2 | 0.9 | 0.989 |
| SMU1 DNA replication regulator and spliceosomal factor | 60695.2 | 0.9 | 0.988 |
| Top 10 interactions for RNY-5 with ORC related proteins | |||
| DNA polymerase eta isoform X2 | 24302234.1 | 0.9 | 0.948 |
| DNA polymerase eta isoform X1 | 5249243.1 | 0.9 | 0.94 |
| APC membrane recruitment protein 3 | 689911.2 | 0.9 | 0.944 |
| DNA ligase 3 isoform beta precursor | 2302.2 | 0.85 | 0.939 |
| DNA repair protein REV1 isoform 6 | 1308389.1 | 0.85 | 0.955 |
| DNA repair protein REV1 isoform 5 | 1308387.1 | 0.85 | 0.956 |
| DNA-directed RNA polymerase III subunit RPC5 isoform 1 | 60589.1 | 0.85 | 0.932 |
| RNA polymerase I-specific transcription initiation factor RRN3 isoform 2 | 1287993.1 | 0.85 | 0.973 |
| Mediator of RNA polymerase II transcription subunit 26 | 4822.2 | 0.85 | 0.929 |
| DNA repair protein REV1 isoform X7 | 24308728.1 | 0.85 | 0.955 |
Table 3: Top 10 interactions with the highest average scores for the interactions of Y-RNAs with the ORC associated proteins.
Protein interactome and PANTHER and string analyses
To gain insight into the biological impact of Y-RNAs interactions with cell receptors, the protein interactome as shown in Figure 2A and the biological processes were analyzed with string and PANTHER. The main biological processes in which these proteins are participating were cellular and metabolic processes, and biological regulation as shown in Figure 3A. Meanwhile, TLRs interactome mainly showed the interaction between TLRs and to a lesser extent with other proteins of with which YRNAs interact as shown in Figure 3B. According to PANTHER, TLRs interactome are involved in the control of metabolic process and response to stimulus as shown in Figure 3B. Origin of Replication Complex (ORC) proteins interactome showed a complex network of interactions mainly involved in biological regulation, cellular process, and developmental process as shown in Figure 3C.
Circulating Y-RNAs showed relative high expression changes among the studied groups and they were predicted to be involved in controlling specific signaling pathways. Their role as biomarkers for p-Ast has to be established with further studies.
This type of RNAs is involved in diverse biological processes, such as cell communication, immune activation and DNA replication [33]. Evidence has demonstrated individual interactions of Y-RNAs with specific proteins as TLRs. However, to our knowledge, Y-RNAs interactions with different protein families have not been tested. Therefore, the present study showed a predictive approach to identify Y-RNAs interactions with different types of proteins. Interactions with TLRs were predicted to validate our bioinformatics model. Data indicate strong interactions of Y-RNAs with protein families not previously described.
The enigma regarding the relative abundance of Y-RNAs and their fragments in serum, as well as their function as circulating molecules it is still unknown. In healthy individuals, circulating Y-RNAs have been described as both free circulating molecules and inside Extracellular Vesicles (EVs), and their overexpression was observed in cancer [34,35]. The predictions of interaction probabilities presented here support the notion that Y-RNAs could be effectively multitasking molecules, regulating key biological processes by controlling metabolism pathways and by affecting cellular processes in cancer or other pathologies. Thus, as an aftermath of molecular interactions with cell receptors or other type of proteins this might result in the immune signaling regulation by activating cellular processes as apoptosis, abnormal cell proliferation, cell cycle alterations, and others. Our predictions showed the interaction of Y-RNAs with diverse cell receptors, which could change cell responses to environment.
The fact that specific subtypes of Y-RNAs showed higher interaction scores with specific proteins could indicate specific functions for each of these ncRNAs. Therefore, this type of studies allows us to predict specific interactions from the whole Y-RNAs molecules or from their fragments with cell receptors, which in turn might trigger signaling cascades depending on the cell receptor repertoire and the relative abundance of the distinct Y-RNAs subtypes in a tissue. The highest score values were observed for RNY-4 and RNY-5 by interacting with different types of proteins. It is of interest to note the high scores for RNY-4 and RNY-5 interactions with TLR10 and TLR5. TLR10, unlike other TLRs, do not activate the immune system and instead they suppress inflammatory signaling [36]. Importantly, TLR10 ligand has not been identified yet and these Y-RNAs could be natural ligands for this receptor. Meanwhile, TLR5 is an immune sensor and a receptor recognizing flagellated bacteria, and it is involved in the control of inflammatory responses and adaptive immunity [37,38]. In innate immunity TLRs induce cell activation leading to cytokine release. Particularly, the activation of TLR3, TLR7, TLR8, and TLR9 induce the expression of inflammatory-cytokines and type I interferons to counteract viral infections. TLRs are ancient elements of the immune system and, similarly to Y-RNAs, they are evolutionarily conserved. Since Human embryonic kidney 293 (HEK 293) cells express TLR-3, TLR-7, or TLR-8, these cells were used to determine the TLR-inducing activities of Y-RNAs. This study showed that human and murine RNY-3 induces the activity of TLR-3, but not that of TLR7, indicating Y-RNAs specificity for TLRs. For the first time, it was demonstrated a differential activation of TLRs by distinct Y-RNA subtypes. According to this study, the authors conclude that Y-RNAs are selective for specific TLRs.
Y-RNAs interactions with the ORC
On average, all four Y-RNAs are significantly overexpressed in solid tumors by 4-fold and 13-fold compared to non-neoplastic tissues. The RNY-1 and RNY-4 expressions were demonstrated. The downregulation of either RNY-1 or RNY-3, or both by siRNAs, leads to a significant reduction of the replicating cells in the S phase, indicating their participation in the Origin of Replication Complex (ORC) formation. In agreement to this, our data showed a strong interaction of RNY-4 with Minichromosome Maintenance Complex Component 3 (MCM3), which is involved in the initiation of the eukaryotic genome replication. This protein, together with other MCM proteins, conform the pre-Replication Complex (pre-RC) and may be involved in the replication fork formation and in the recruitment of other DNA replication of related proteins [39]. In pediatric patients with Ast, the circulating RNY-4 expression was considerably diminished suggesting a loss of its regulatory function in comparison to controls; however, it is important to know if the concentration of free or packed RNY-4 in other Extracellular Vesicles (Evs) also decreases. By contrast, the RNY-5 expression was upregulated, but its interaction scores were not so significant. The identification of the target cells of the exosomes pediatric patients with Ast is crucial, since this will make it possible to know if RNY-4 is integrated to the Origin of Replication Complex (ORC) of these cells or have different functions.
Y-RNAs loop domain
An interesting result was the fact that the Y-RNAs loop domain by itself interacted with TLRs, supporting the notion that processed Y-RNA fragments could be functional [40]. This domain is the least conserved in Y-RNAs and, according to this, it is speculated that the loop domain gives a specific function to each Y-RNA among species. In fact, this domain could be participating in specific cellular functions [41,42]. Either as activator or modulator of their function and has been experimentally used for several protein binding sites for the Y-RNAs loop domain for proteins such as Nucleolin (NCL), Polypyrimidine-tract Binding Protein 1 (PTBP1) and Z-DNA-Binding Protein 1 (ZBP1) [9].
This study generated new questions without answers: What are the target tissues of these RNAs? What are their functions? Does the function of circulating Y-RNAs depend on whether they travel freely through the bloodstream or they go inside Extracellular Vesicles (EVs)? Y-RNAs might be involved in many cellular functions, depending on many factors, such as the developmental stage of the organism, the cell type in which each Y-RNA is expressed, the combination of Y-RNAs expressed in the same cell type, and the proteins and other RNAs that are co-expressed with them, among others. Our data showed that Y-RNAs could be interacting with many types of proteins to regulate specific cell responses, but experimental verification is still necessary. Despite the above, Y-RNAs shown to be potential biomarkers for LgAst diagnosis and prognosis.
JMRC was supported by the Consejo Nacional de Ciencias y Tecnología (CONACyT) with a doctoral grant scholarship: CVU: 101863 Support: 472835. The authors thank Research Square the Pre-print. We thank Sintagma Translations for the English proofreading and editing.
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
Citation: Rodríguez-Corona JM, Esparza-Garrido RR, Horta Vega JV, Velazquez Flores MA (2023) Circulating Y-RNAs: A Predicted Function Mainly in Controlling the Innate Immune System, Cell Signaling, and DNA Replication in Pediatric Patients with Pilocytic and Diffuse Astrocytoma. Clin Pediatr. 8:239.
Received: 01-May-2023, Manuscript No. CPOA-23-23818 ; Editor assigned: 03-May-2023, Pre QC No. CPOA-23-23818 (PQ); Reviewed: 17-May-2023, QC No. CPOA-23-23818 ; Revised: 24-May-2023, Manuscript No. CPOA-23-23818 (R); Published: 02-Jun-2023 , DOI: 10.35248/2572-0775.23.8.239
Copyright: © 2023 Rodríguez-Corona JM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.